Diet and microbiome in the beginning of the sequence of gut inflammation
- PMID: 35071544
- PMCID: PMC8717522
- DOI: 10.12998/wjcc.v9.i36.11122
Diet and microbiome in the beginning of the sequence of gut inflammation
Abstract
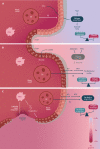
Inflammatory bowel disease (IBD) is a chronic inflammatory condition of the gastrointestinal tract due, at least partially, to an aberrant and excessive mucosal immune response to gut bacteria in genetically-predisposed individuals under certain environmental factors. The incidence of IBD is rising in western and newly industrialized countries, paralleling the increase of westernized dietary patterns, through new antigens, epithelial function and permeability, epigenetic mechanisms (e.g., DNA methylation), and alteration of the gut microbiome. Alteration in the composition and functionality of the gut microbiome (including bacteria, viruses and fungi) seems to be a nuclear pathogenic factor. The microbiome itself is dynamic, and the changes in food quality, dietary habits, living conditions and hygiene of these western societies, could interact in a complex manner as modulators of dysbiosis, thereby influencing the activation of immune cells' promoting inflammation. The microbiome produces diverse small molecules via several metabolic ways, with the fiber-derived short-chain fatty acids (i.e., butyrate) as main elements and having anti-inflammatory effects. These metabolites and some micronutrients of the diet (i.e., vitamins, folic acid, beta carotene and trace elements) are regulators of innate and adaptive intestinal immune homeostasis. An excessive and unhealthy consumption of sugar, animal fat and a low-vegetable and -fiber diet are risk factors for IBD appearance. Furthermore, metabolism of nutrients in intestinal epithelium and in gut microbiota is altered by inflammation, changing the demand for nutrients needed for homeostasis. This role of food and a reduced gut microbial diversity in causing IBD might also have a prophylactic or therapeutic role for IBD. The relationship between dietary intake, symptoms, and bowel inflammation could lead to dietary and lifestyle recommendations, including diets with abundant fruits, vegetables, olive oil and oily fish, which have anti-inflammatory effects and could prevent dysbiosis and IBD. Dietary modulation and appropriate exclusion diets might be a new complementary management for treatment at disease flares and in refractory patients, even reducing complications, hospitalizations and surgery, through modifying the luminal intestinal environment.
Keywords: Diet; Inflammatory bowel disease; Microbiome; Pathogenia.
©The Author(s) 2021. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors declare no conflicts of interests.
Figures

References
-
- Nataro JP, Cohen PS, Mobley HLT, Weiser JN. Colonization of mucosal surfaces. American Society for Microbiology, 2005.
-
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. - PubMed
-
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

