Sequence-selective purification of biological RNAs using DNA nanoswitches
- PMID: 35072148
- PMCID: PMC8782281
- DOI: 10.1016/j.crmeth.2021.100126
Sequence-selective purification of biological RNAs using DNA nanoswitches
Abstract
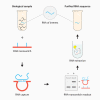
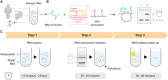
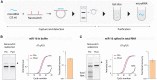
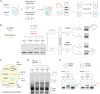
Nucleic acid purification is a critical aspect of biomedical research and a multibillion-dollar industry. Here we establish sequence-selective RNA capture, release, and isolation using conformationally responsive DNA nanoswitches. We validate purification of specific RNAs ranging in size from 22 to 401 nt with up to 75% recovery and 99.98% purity in a benchtop process with minimal expense and equipment. Our method compared favorably with bead-based extraction of an endogenous microRNA from cellular total RNA, and can be programmed for multiplexed purification of multiple individual RNA targets from one sample. Coupling our approach with downstream LC/MS, we analyzed RNA modifications in 5.8S ribosomal RNA, and found 2'-O-methylguanosine, 2'-O-methyluridine, and pseudouridine in a ratio of ~1:7:22. The simplicity, low cost, and low sample requirements of our method make it suitable for easy adoption, and the versatility of the approach provides opportunities to expand the strategy to other biomolecules.
Conflict of interest statement
DECLARATION OF INTERESTS K.H. and A.R.C. have intellectual property related to DNA nanoswitches. L.Z. and K.H. are inventors on a patent application covering aspects of this work. All other authors declare that they have no competing interests.
Figures


Similar articles
-
Single species RNA purification using DNA nanoswitches.Trends Biochem Sci. 2022 Apr;47(4):367-368. doi: 10.1016/j.tibs.2021.12.006. Epub 2022 Jan 10. Trends Biochem Sci. 2022. PMID: 35027255 Free PMC article.
-
Multiplexed Nanopore-Based Nucleic Acid Sensing and Bacterial Identification Using DNA Dumbbell Nanoswitches.J Am Chem Soc. 2023 Jun 7;145(22):12115-12123. doi: 10.1021/jacs.3c01649. Epub 2023 May 23. J Am Chem Soc. 2023. PMID: 37220424 Free PMC article.
-
How to Perform miRacles: A Step-by-Step microRNA Detection Protocol Using DNA Nanoswitches.Curr Protoc Mol Biol. 2020 Mar;130(1):e114. doi: 10.1002/cpmb.114. Curr Protoc Mol Biol. 2020. PMID: 32048806 Free PMC article.
-
Identification of modified residues in RNAs by reverse transcription-based methods.Methods Enzymol. 2007;425:21-53. doi: 10.1016/S0076-6879(07)25002-5. Methods Enzymol. 2007. PMID: 17673078 Review.
-
Pseudouridine and O2'-methylated nucleosides. Significance of their selective occurrence in rRNA domains that function in ribosome-catalyzed synthesis of the peptide bonds in proteins.Biochimie. 1995;77(1-2):7-15. doi: 10.1016/0300-9084(96)88098-9. Biochimie. 1995. PMID: 7599278 Review.
Cited by
-
Heteromultivalency enables enhanced detection of nucleic acid mutations.Nat Chem. 2024 Feb;16(2):229-238. doi: 10.1038/s41557-023-01345-4. Epub 2023 Oct 26. Nat Chem. 2024. PMID: 37884668
-
A novel method to purify small RNAs from human tissues for methylation analysis by LC-MS/MS.Front Mol Biosci. 2022 Aug 30;9:949181. doi: 10.3389/fmolb.2022.949181. eCollection 2022. Front Mol Biosci. 2022. PMID: 36111135 Free PMC article.
-
Single species RNA purification using DNA nanoswitches.Trends Biochem Sci. 2022 Apr;47(4):367-368. doi: 10.1016/j.tibs.2021.12.006. Epub 2022 Jan 10. Trends Biochem Sci. 2022. PMID: 35027255 Free PMC article.
-
Prevention of ribozyme catalysis through cDNA synthesis enables accurate RT-qPCR measurements of context-dependent ribozyme activity.RNA. 2025 Apr 16;31(5):633-645. doi: 10.1261/rna.080243.124. RNA. 2025. PMID: 40050070
-
Multiplexed Nanopore-Based Nucleic Acid Sensing and Bacterial Identification Using DNA Dumbbell Nanoswitches.J Am Chem Soc. 2023 Jun 7;145(22):12115-12123. doi: 10.1021/jacs.3c01649. Epub 2023 May 23. J Am Chem Soc. 2023. PMID: 37220424 Free PMC article.
References
-
- Adams N.M., Bordelon H., Wang K.-K.A., Albert L.E., Wright D.W., Haselton F.R. Comparison of three magnetic bead surface functionalities for RNA extraction and detection. ACS Appl. Mater. Inter. 2015;7:6062–6069. - PubMed
-
- Aqeilan R.I., Calin G.A., Croce C.M. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

