Effects of Wood Smoke Constituents on Mucin Gene Expression in Mice and Human Airway Epithelial Cells and on Nasal Epithelia of Subjects with a Susceptibility Gene Variant in Tp53
- PMID: 35072516
- PMCID: PMC8785869
- DOI: 10.1289/EHP9446
Effects of Wood Smoke Constituents on Mucin Gene Expression in Mice and Human Airway Epithelial Cells and on Nasal Epithelia of Subjects with a Susceptibility Gene Variant in Tp53
Abstract
Background: Exposure to wood smoke (WS) increases the risk for chronic bronchitis more than exposure to cigarette smoke (CS), but the underlying mechanisms are unclear.
Objective: The effect of WS and CS on mucous cell hyperplasia in mice and in human primary airway epithelial cells (AECs) was compared with replicate the findings in human cohorts. Responsible WS constituents were identified to better delineate the pathway involved, and the role of a tumor protein p53 (Tp53) gene polymorphism was investigated.
Methods: Mice and primary human AECs were exposed to WS or CS and the signaling receptor and pathway were identified using short hairpin structures, small molecule inhibitors, and Western analyses. Mass spectrometric analysis was used to identify active WS constituents. The role of a gene variant in Tp53 that modifies proline to arginine was examined using nasal brushings from study participants in the Lovelace Smokers Cohort, primary human AECs, and mice with a modified Tp53 gene.
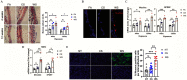
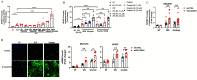
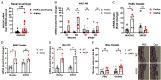
Results: WS at 25-fold lower concentration than CS increased mucin expression more efficiently in mice and in human AECs in a p53 pathway-dependent manner. Study participants who were homozygous for p53 arginine compared with the proline variant showed higher mucin 5AC (MUC5AC) mRNA levels in nasal brushings if they reported WS exposure. The WS constituent, oxalate, increased MUC5AC levels similar to the whole WS extract, especially in primary human AECs homozygous for p53 arginine, and in mice with a modified Tp53 gene. Further, the anion exchange protein, SLC26A9, when reduced, enhanced WS- and oxalate-induced mucin expression.
Discussion: The potency of WS compared with CS in inducing mucin expression may explain the increased risk for chronic bronchitis in participants exposed to WS. Identification of the responsible compounds could help estimate the risk of pollutants in causing chronic bronchitis in susceptible individuals and provide strategies to improve management of lung diseases. https://doi.org/10.1289/EHP9446.
Figures


References
-
- Allinson JP, Hardy R, Donaldson GC, Shaheen SO, Kuh D, Wedzicha JA. 2016. The presence of chronic mucus hypersecretion across adult life in relation to chronic obstructive pulmonary disease development. Am J Respir Crit Care Med 193(6):662–672, PMID: , 10.1164/rccm.201511-2210OC. - DOI - PMC - PubMed
-
- Awji EG, Chand H, Bruse S, Smith KR, Colby JK, Mebratu Y, et al. 2015. Wood smoke enhances cigarette smoke-induced inflammation by inducing the aryl hydrocarbon receptor repressor in airway epithelial cells. Am J Respir Cell Mol Biol 52(3):377–386, PMID: , 10.1165/rcmb.2014-0142OC. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

