ROSIE: RObust Sparse ensemble for outlIEr detection and gene selection in cancer omics data
- PMID: 35072570
- PMCID: PMC9014683
- DOI: 10.1177/09622802211072456
ROSIE: RObust Sparse ensemble for outlIEr detection and gene selection in cancer omics data
Abstract
The extraction of novel information from omics data is a challenging task, in particular, since the number of features (e.g. genes) often far exceeds the number of samples. In such a setting, conventional parameter estimation leads to ill-posed optimization problems, and regularization may be required. In addition, outliers can largely impact classification accuracy.Here we introduce ROSIE, an ensemble classification approach, which combines three sparse and robust classification methods for outlier detection and feature selection and further performs a bootstrap-based validity check. Outliers of ROSIE are determined by the rank product test using outlier rankings of all three methods, and important features are selected as features commonly selected by all methods.We apply ROSIE to RNA-Seq data from The Cancer Genome Atlas (TCGA) to classify observations into Triple-Negative Breast Cancer (TNBC) and non-TNBC tissue samples. The pre-processed dataset consists of genes and more than samples. We demonstrate that ROSIE selects important features and outliers in a robust way. Identified outliers are concordant with the distribution of the commonly selected genes by the three methods, and results are in line with other independent studies. Furthermore, we discuss the association of some of the selected genes with the TNBC subtype in other investigations. In summary, ROSIE constitutes a robust and sparse procedure to identify outliers and important genes through binary classification. Our approach is ad hoc applicable to other datasets, fulfilling the overall goal of simultaneously identifying outliers and candidate disease biomarkers to the targeted in therapy research and personalized medicine frameworks.
Keywords: Ensemble; biomarker; classification; feature selection; outlier; robust; sparse; triple-Negative Breast Cancer.
Conflict of interest statement
Figures
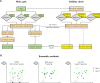
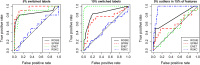
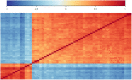
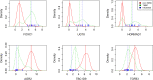

References
-
- Thomas RS, Wesselkamper SC, Wang NCY. et al.. Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol Sci 2013; 134: 180–194. DOI: 10.1093/toxsci/kft094. https://academic.oup.com/toxsci/article-pdf/134/1/180/16685755/kft094.pdf . - DOI - PubMed
-
- Sutherland J, Webster Y, Willy J. et al.. Toxicogenomic module associations with pathogenesis: a network-based approach to understanding drug toxicity. Pharmacogenomics J 2018; 18: 377–390. - PubMed
-
- Zhang X, Yap Y, Wei D. et al.. Novel omics technologies in nutrition research. Biotechnol Adv 2008; 26: 169–176. DOI: 10.1016/j.biotechadv.2007.11.002. https://www.sciencedirect.com/science/article/pii/S0734975007001206 . - DOI - PubMed
-
- Kato H, Takahashi S, Saito K. Omics and integrated omics for the promotion of food and nutrition science. J Tradit Complement Med 2011; 1: 25–30. DOI: 10.1016/S2225-4110(16)30053-0. https://www.sciencedirect.com/science/article/pii/S2225411016300530 . - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

