Bacterial Indole as a Multifunctional Regulator of Klebsiella oxytoca Complex Enterotoxicity
- PMID: 35073747
- PMCID: PMC8787480
- DOI: 10.1128/mbio.03752-21
Bacterial Indole as a Multifunctional Regulator of Klebsiella oxytoca Complex Enterotoxicity
Abstract
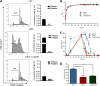
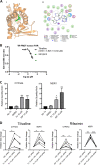
Gastrointestinal microbes respond to biochemical metabolites that coordinate their behaviors. Here, we demonstrate that bacterial indole functions as a multifactorial mitigator of Klebsiella grimontii and Klebsiella oxytoca pathogenicity. These closely related microbes produce the enterotoxins tilimycin and tilivalline; cytotoxin-producing strains are the causative agent of antibiotic-associated hemorrhagic colitis and have been associated with necrotizing enterocolitis of premature infants. We demonstrate that carbohydrates induce cytotoxin synthesis while concurrently repressing indole biosynthesis. Conversely, indole represses cytotoxin production. In both cases, the alterations stemmed from differential transcription of npsA and npsB, key genes involved in tilimycin biosynthesis. Indole also enhances conversion of tilimycin to tilivalline, an indole analog with reduced cytotoxicity. In this context, we established that tilivalline, but not tilimycin, is a strong agonist of pregnane X receptor (PXR), a master regulator of xenobiotic detoxification and intestinal inflammation. Tilivalline binding upregulated PXR-responsive detoxifying genes and inhibited tubulin-directed toxicity. Bacterial indole, therefore, acts in a multifunctional manner to mitigate cytotoxicity by Klebsiella spp.: suppression of toxin production, enhanced conversion of tilimycin to tilivalline, and activation of PXR. IMPORTANCE The human gut harbors a complex community of microbes, including several species and strains that could be commensals or pathogens depending on context. The specific environmental conditions under which a resident microbe changes its relationship with a host and adopts pathogenic behaviors, in many cases, remain poorly understood. Here, we describe a novel communication network involving the regulation of K. grimontii and K. oxytoca enterotoxicity. Bacterial indole was identified as a central modulator of these colitogenic microbes by suppressing bacterial toxin (tilimycin) synthesis and converting tilimycin to tilivalline while simultaneously activating a host receptor, PXR, as a means of mitigating tissue cytotoxicity. On the other hand, fermentable carbohydrates were found to inhibit indole biosynthesis and enhance toxin production. This integrated network involving microbial, host, and metabolic factors provides a contextual framework to better understand K. oxytoca complex pathogenicity.
Keywords: Klebsiella oxytoca complex; cytotoxin; indole; intestinal inflammation; pregnane X receptor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
