N7-Methylation of the Coronavirus RNA Cap Is Required for Maximal Virulence by Preventing Innate Immune Recognition
- PMID: 35073761
- PMCID: PMC8787479
- DOI: 10.1128/mbio.03662-21
N7-Methylation of the Coronavirus RNA Cap Is Required for Maximal Virulence by Preventing Innate Immune Recognition
Abstract
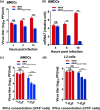
The ongoing coronavirus (CoV) disease 2019 (COVID-19) pandemic caused by infection with severe acute respiratory syndrome CoV 2 (SARS-CoV-2) is associated with substantial morbidity and mortality. Understanding the immunological and pathological processes of coronavirus diseases is crucial for the rational design of effective vaccines and therapies for COVID-19. Previous studies showed that 2'-O-methylation of the viral RNA cap structure is required to prevent the recognition of viral RNAs by intracellular innate sensors. Here, we demonstrate that the guanine N7-methylation of the 5' cap mediated by coronavirus nonstructural protein 14 (nsp14) contributes to viral evasion of the type I interferon (IFN-I)-mediated immune response and pathogenesis in mice. A Y414A substitution in nsp14 of the coronavirus mouse hepatitis virus (MHV) significantly decreased N7-methyltransferase activity and reduced guanine N7-methylation of the 5' cap in vitro. Infection of myeloid cells with recombinant MHV harboring the nsp14-Y414A mutation (rMHVnsp14-Y414A) resulted in upregulated expression of IFN-I and ISG15 mainly via MDA5 signaling and in reduced viral replication compared to that of wild-type rMHV. rMHVnsp14-Y414A replicated to lower titers in livers and brains and exhibited an attenuated phenotype in mice. This attenuated phenotype was IFN-I dependent because the virulence of the rMHVnsp14-Y414A mutant was restored in Ifnar-/- mice. We further found that the comparable mutation (Y420A) in SARS-CoV-2 nsp14 (rSARS-CoV-2nsp14-Y420A) also significantly decreased N7-methyltransferase activity in vitro, and the mutant virus was attenuated in K18-human ACE2 transgenic mice. Moreover, infection with rSARS-CoV-2nsp14-Y420A conferred complete protection against subsequent and otherwise lethal SARS-CoV-2 infection in mice, indicating the vaccine potential of this mutant. IMPORTANCE Coronaviruses (CoVs), including SARS-CoV-2, the cause of COVID-19, use several strategies to evade the host innate immune responses. While the cap structure of RNA, including CoV RNA, is important for translation, previous studies indicate that the cap also contributes to viral evasion from the host immune response. In this study, we demonstrate that the N7-methylated cap structure of CoV RNA is pivotal for virus immunoevasion. Using recombinant MHV and SARS-CoV-2 encoding an inactive N7-methyltransferase, we demonstrate that these mutant viruses are highly attenuated in vivo and that attenuation is apparent at very early times after infection. Virulence is restored in mice lacking interferon signaling. Further, we show that infection with virus defective in N7-methylation protects mice from lethal SARS-CoV-2, suggesting that the N7-methylase might be a useful target in drug and vaccine development.
Keywords: N7-methylation; RNA cap structure; RNA methylation; SARS-CoV-2; coronavirus; immune response; interferons; nonstructural protein 14 (nsp14); type I interferon.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr, Shi PY, Diamond MS. 2010. 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi:10.1038/nature09489. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
