Genomic diversity and antimicrobial resistance of Campylobacter spp. from humans and livestock in Nigeria
- PMID: 35073916
- PMCID: PMC8788075
- DOI: 10.1186/s12929-022-00786-2
Genomic diversity and antimicrobial resistance of Campylobacter spp. from humans and livestock in Nigeria
Abstract
Background: Campylobacter spp. are zoonotic pathogens, ubiquitous and are found naturally as commensals in livestock from where they can be transmitted to humans directly or through animal products. The genomic diversity and antimicrobial resistance profile of Campylobacter was investigated with a focus on C. jejuni and C. coli in humans and livestock (poultry and cattle) from Nigeria.
Methods: 586 human stool samples and 472 faecal samples from livestock were cultured for thermophilic Campylobacter species on modified charcoal cefoperazone deoxycholate agar (mCCDA). Culture in combination with whole genome sequencing identified and confirmed the presence of Campylobacter in humans and animals from the study area. Further analysis of the sequences was performed to determine multilocus sequence types and genetic determinants of antimicrobial resistance to fluoroquinolone, betalactam, tetracycline and macrolide classes of antimicrobials.
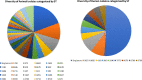
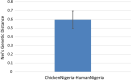
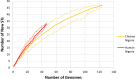
Results: From the human stool samples tested, 50 (9%) were positive of which 33 (66%) were C. jejuni, 14 (28%) were C. coli while 3 (6%) were C. hyointestinalis. In livestock, 132 (28%) were positive. Thirty one (7%) were C. jejuni while 101 (21%) were C. coli. Whole genome sequencing and MLST of the isolates revealed a total of 32 sequence types (STs) identified from 47 human isolates while 48 STs were identified in 124 isolates from livestock indicating a population which was overall, genetically diverse with a few more dominant strains. The antimicrobial resistance profiles of the isolates indicated a higher prevalence of resistance in Campylobacter isolated from livestock than in humans. Generally, resistance was greatest for betalactams (42%) closely followed by fluoroquinolones (41%), tetracyclines (15%) and lastly macrolides (2%). Multidrug resistance to three or more antimicrobials was observed in 24 (13%) isolates from humans (n = 1, 4%) and chicken (n = 23, 96%).
Conclusions: This study has further contributed information about the epidemiology, genetic diversity and antimicrobial resistance profile of thermophilic Campylobacter in Nigeria.
Keywords: Antimicrobial resistance; Campylobacter; Clonal complex; Multilocus sequence typing; Sequence type.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375(9730):1969–1987. - PubMed
-
- Baqar S, Bourgeois AL, Schultheiss PJ, Walker RI, Rollins DM, Haberberger RL, et al. Safety and immunogenicity of a prototype oral whole-cell killed Campylobacter vaccine administered with a mucosal adjuvant in non-human primates. Vaccine. 1995;13(1):22–28. - PubMed
-
- Rao MR, Naficy AB, Savarino SJ, Abu-Elyazeed R, Wierzba TF, Peruski LF, et al. Pathogenicity and convalescent excretion of Campylobacter in rural Egyptian children. Am J Epidemiol. 2001;154(2):166–173. - PubMed
-
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403(6770):665–668. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

