Identification of genetic risk loci and prioritization of genes and pathways for myasthenia gravis: a genome-wide association study
- PMID: 35074870
- PMCID: PMC8812681
- DOI: 10.1073/pnas.2108672119
Identification of genetic risk loci and prioritization of genes and pathways for myasthenia gravis: a genome-wide association study
Erratum in
-
Correction for Chia et al., Identification of genetic risk loci and prioritization of genes and pathways for myasthenia gravis: a genome-wide association study.Proc Natl Acad Sci U S A. 2022 Jun 7;119(23):e2206754119. doi: 10.1073/pnas.2206754119. Epub 2022 Jun 3. Proc Natl Acad Sci U S A. 2022. PMID: 35658084 Free PMC article. No abstract available.
Abstract
Myasthenia gravis is a chronic autoimmune disease characterized by autoantibody-mediated interference of signal transmission across the neuromuscular junction. We performed a genome-wide association study (GWAS) involving 1,873 patients diagnosed with acetylcholine receptor antibody-positive myasthenia gravis and 36,370 healthy individuals to identify disease-associated genetic risk loci. Replication of the discovered loci was attempted in an independent cohort from the UK Biobank. We also performed a transcriptome-wide association study (TWAS) using expression data from skeletal muscle, whole blood, and tibial nerve to test the effects of disease-associated polymorphisms on gene expression. We discovered two signals in the genes encoding acetylcholine receptor subunits that are the most common antigenic target of the autoantibodies: a GWAS signal within the cholinergic receptor nicotinic alpha 1 subunit (CHRNA1) gene and a TWAS association with the cholinergic receptor nicotinic beta 1 subunit (CHRNB1) gene in normal skeletal muscle. Two other loci were discovered on 10p14 and 11q21, and the previous association signals at PTPN22, HLA-DQA1/HLA-B, and TNFRSF11A were confirmed. Subgroup analyses demonstrate that early- and late-onset cases have different genetic risk factors. Genetic correlation analysis confirmed a genetic link between myasthenia gravis and other autoimmune diseases, such as hypothyroidism, rheumatoid arthritis, multiple sclerosis, and type 1 diabetes. Finally, we applied Priority Index analysis to identify potentially druggable genes/proteins and pathways. This study provides insight into the genetic architecture underlying myasthenia gravis and demonstrates that genetic factors within the loci encoding acetylcholine receptor subunits contribute to its pathogenesis.
Keywords: genetic correlation; genome-wide association study; myasthenia gravis; pathway analysis.
Copyright © 2022 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: R.J.B. served as a consultant for NuFactor and Momenta Pharmaceutical and receives research support from PTC Therapeutics, Ra Pharma, Orphazyme, Sanofi Genzyme, FDA OOPD, NIH, and Patient-Centered Outcomes Research Institute (PCORI). M.B. reports grant support from Muscular Dystrophy Association, ALS Association, ALS Recovery Fund, Kimmelman Estate, Target ALS, Eli Lilly and Company, and NIH during the conduct of the study. He also reports grant support from FDA, Centers for Disease Control and Prevention (CDC), and DOD; research support from Alexion Pharmaceuticals, UCB, Cytokinetics, Neuraltus, Biogen, and Orphazyme A/S; and personal fees from NMD Pharma, Ra Pharmaceuticals, Mitsubishi Tanabe, Avexis, UCB, and Denali outside the submitted work. V.C. served as a consultant for review and expert testimony for the Department of Health and Human Services and the Department of Justice under the Vaccine Injury and Compensation Program. Dr. Chaudhry has received a royalty for total neuropathy score (TNS) patented (through Johns Hopkins University) for the license of TNS use from AstraZeneca, Genentech, Seattle Genetics, Calithera Biosciences, Merrimack Pharmaceuticals, Levicept, and Acetylon Pharmaceuticals. M.M.D. serves or recently served as a consultant for Argenx Pharmaceuticals, Catalyst, CSL Behring, Kezar, Momenta, NuFactor, RMS Medical, Sanofi Genzyme, Shire Takeda, and Spark Therapeutics. Dr. Dimachkie received grants from Alexion Pharmaceuticals, Alnylam Pharmaceuticals, Amicus, Biomarin, Bristol-Myers Squibb, Catalyst, CSL Behring, FDA/OOPD, GlaxoSmithKline, Genentech, Grifols, Mitsubishi Tanabe Pharma, Muscular Dystrophy Association (MDA), NIH, Novartis, Sanofi Genzyme, Octapharma, Orphazyme, Sarepta Therapeutics, Shire Takeda, Spark, UCB Biopharma, Viromed, and TMA. A.E. was a member of the advisory board for Alexion Pharmaceuticals, a scientific award jury member for Grifols, and safety data monitor for UCB. M.F. has received honoraria for serving on advisory boards for Argenx Pharmaceuticals and Alexion Pharmaceuticals. M.F. also has research support from Catalyst, Ra pharma, Amicus, Orphazyme, Alexion Pharmaceuticals, Momenta, and Alnylam. J.F.H. reports research support and grants from Alexion Pharmaceuticals and Argenx Pharmaceuticals, Centers for Disease Control and Prevention, MDA, NIH (including the National Institute of Neurologic Disorders and Stroke and the National Institute of Arthritis and Musculoskeletal and Skin Disease), PCORI, and Ra Pharmaceuticals (now UCB Biosciences); honoraria from Alexion Pharmaceuticals, Argenx Pharmaceuticals, Immunovant, Ra Pharmaceuticals (now UCB Biosciences), Regeneron Pharmaceuticals, and Viela Bio; and nonfinancial support from Alexion Pharmaceuticals, Argenx Pharmaceuticals, Ra Pharmaceuticals (now UCB Biosciences), and Toleranzia. P.J.T. holds patents on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72 and has received grant funding from the Helsinki University Hospital, Finnish Academy, Sigrid Juselius Foundation, EU Marie Curie action, Biogen Finland, Roche Finland, Merck Finland, Sanofi Genzyme Finland, and Novartis Finland and has made consultations to Alexion Pharmaceuticals, Biogen, Novartis, Orion, Roche, Sanofi Genzyme, Santen, and Teva. H.J.K. is funded by the MDA (508240) and NIH grant U54NS115054; is a consultant for Alnylam Pharmaceuticals, Ra Pharmaceuticals, and UCB Pharmaceuticals; and is CEO of ARC Biotechnology, LLC, which receives support from the NIH (R41NS110331). He serves on the Editorial Board of Experimental Neurology. M.M.M. has received honoraria as a speaker and/or moderator from Alnylam, Akcea, Pfizer, and CSL Behring. She has served on Advisory Boards for Pfizer, Alnylam, and Akcea. She serves as an investigator for clinical trials with Alnylam and Biogen. S.M. has served on advisory board meetings for Alexion Pharmaceuticals and Argenx Pharmaceuticals. M.P. served on the advisory boards for CSL Behring, Alexion Pharmaceuticals, and Argenx Pharmaceuticals and has been a consultant for Momenta Pharmaceuticals. D.P.R. receives research funding from a Sponsored Research Agreement from Cabaletta Bioscience. B.J.T. holds patents on the clinical testing and therapeutic intervention for the hexanucleotide repeat expansion of C9orf72 and has received research grants from the Myasthenia Gravis Foundation, the Robert Packard Center for ALS Research, the ALS Association, the Italian Football Federation, the CDC, the MDA, Merck, and Microsoft Research. B.J.T. receives funding through the Intramural Research Program at NIH.
Figures

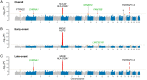


Comment in
-
Reply to Zhu et al.: Implications of CHRNB1 and ERBB2 in the pathobiology of myasthenia gravis.Proc Natl Acad Sci U S A. 2022 Sep 9;119(36):e2209096119. doi: 10.1073/pnas.2209096119. Epub 2022 Aug 15. Proc Natl Acad Sci U S A. 2022. PMID: 35969799 Free PMC article. No abstract available.
-
Novel genes/loci validate the small effect size of ERBB2 in patients with myasthenia gravis.Proc Natl Acad Sci U S A. 2022 Sep 9;119(36):e2207273119. doi: 10.1073/pnas.2207273119. Epub 2022 Aug 15. Proc Natl Acad Sci U S A. 2022. PMID: 35969801 Free PMC article. No abstract available.
References
-
- Alshekhlee A., Miles J. D., Katirji B., Preston D. C., Kaminski H. J., Incidence and mortality rates of myasthenia gravis and myasthenic crisis in US hospitals. Neurology 72, 1548–1554 (2009). - PubMed
-
- Vincent A., Palace J., Hilton-Jones D., Myasthenia gravis. Lancet 357, 2122–2128 (2001). - PubMed
-
- Gilhus N. E., Myasthenia Gravis. N. Engl. J. Med. 375, 2570–2581 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 EY016514/EY/NEI NIH HHS/United States
- K05 CA124911/CA/NCI NIH HHS/United States
- U01 HG004446/HG/NHGRI NIH HHS/United States
- P50 CA097007/CA/NCI NIH HHS/United States
- R01 ES011740/ES/NIEHS NIH HHS/United States
- R01 HL076784/HL/NHLBI NIH HHS/United States
- P50 DA019706/DA/NIDA NIH HHS/United States
- R01 CA136725/CA/NCI NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- HHSN268201600004C/HL/NHLBI NIH HHS/United States
- R37 HL039693/HL/NHLBI NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- Z01 AG000949/ImNIH/Intramural NIH HHS/United States
- R01 CA133996/CA/NCI NIH HHS/United States
- HHSN268201600002C/HL/NHLBI NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- HHSN268201600018C/HL/NHLBI NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- R01 EY016482/EY/NEI NIH HHS/United States
- R01 DA013324/DA/NIDA NIH HHS/United States
- P01 CA089392/CA/NCI NIH HHS/United States
- R41 NS110331/NS/NINDS NIH HHS/United States
- U01 HG004424/HG/NHGRI NIH HHS/United States
- R01 DA009532/DA/NIDA NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- P50 CA084724/CA/NCI NIH HHS/United States
- R01 DA026141/DA/NIDA NIH HHS/United States
- U01 HL053941/HL/NHLBI NIH HHS/United States
- HHSN268201600003C/HL/NHLBI NIH HHS/United States
- U01 HG005157/HG/NHGRI NIH HHS/United States
- HHSN268201600001C/HL/NHLBI NIH HHS/United States
- U01 HG005152/HG/NHGRI NIH HHS/United States
- R01 HL070100/HL/NHLBI NIH HHS/United States
- P50 CA093459/CA/NCI NIH HHS/United States
- U54 NS115054/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

