Impact of hemolysis on multi-OMIC pancreatic biomarker discovery to derisk biomarker development in precision medicine studies
- PMID: 35075163
- PMCID: PMC8786830
- DOI: 10.1038/s41598-022-05152-8
Impact of hemolysis on multi-OMIC pancreatic biomarker discovery to derisk biomarker development in precision medicine studies
Abstract
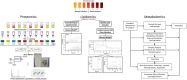
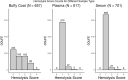
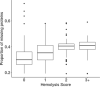
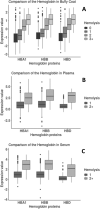
Cancer biomarker discovery is critically dependent on the integrity of biofluid and tissue samples acquired from study participants. Multi-omic profiling of candidate protein, lipid, and metabolite biomarkers is confounded by timing and fasting status of sample collection, participant demographics and treatment exposures of the study population. Contamination by hemoglobin, whether caused by hemolysis during sample preparation or underlying red cell fragility, contributes 0-10 g/L of extraneous protein to plasma, serum, and Buffy coat samples and may interfere with biomarker detection and validation. We analyzed 617 plasma, 701 serum, and 657 buffy coat samples from a 7-year longitudinal multi-omic biomarker discovery program evaluating 400+ participants with or at risk for pancreatic cancer, known as Project Survival. Hemolysis was undetectable in 93.1% of plasma and 95.0% of serum samples, whereas only 37.1% of buffy coat samples were free of contamination by hemoglobin. Regression analysis of multi-omic data demonstrated a statistically significant correlation between hemoglobin concentration and the resulting pattern of analyte detection and concentration. Although hemolysis had the greatest impact on identification and quantitation of the proteome, distinct differentials in metabolomics and lipidomics were also observed and correlated with severity. We conclude that quality control is vital to accurate detection of informative molecular differentials using OMIC technologies and that caution must be exercised to minimize the impact of hemolysis as a factor driving false discovery in large cancer biomarker studies.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Mescher AL. Junqueira's basic histology: text and atlas. New York: McGraw-Hill; 2013.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

