Morphological, physiological, biochemical, and transcriptome studies reveal the importance of transporters and stress signaling pathways during salinity stress in Prunus
- PMID: 35075204
- PMCID: PMC8786923
- DOI: 10.1038/s41598-022-05202-1
Morphological, physiological, biochemical, and transcriptome studies reveal the importance of transporters and stress signaling pathways during salinity stress in Prunus
Abstract
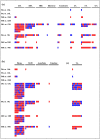
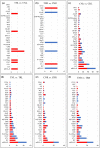
The almond crop has high economic importance on a global scale, but its sensitivity to salinity stress can cause severe yield losses. Salt-tolerant rootstocks are vital for crop economic feasibility under saline conditions. Two commercial rootstocks submitted to salinity, and evaluated through different parameters, had contrasting results with the survival rates of 90.6% for 'Rootpac 40' (tolerant) and 38.9% for 'Nemaguard' (sensitive) under salinity (Electrical conductivity of water = 3 dS m-1). Under salinity, 'Rootpac 40' accumulated less Na and Cl and more K in leaves than 'Nemaguard'. Increased proline accumulation in 'Nemaguard' indicated that it was highly stressed by salinity compared to 'Rootpac 40'. RNA-Seq analysis revealed that a higher degree of differential gene expression was controlled by genotype rather than by treatment. Differentially expressed genes (DEGs) provided insight into the regulation of salinity tolerance in Prunus. DEGs associated with stress signaling pathways and transporters may play essential roles in the salinity tolerance of Prunus. Some additional vital players involved in salinity stress in Prunus include CBL10, AKT1, KUP8, Prupe.3G053200 (chloride channel), and Prupe.7G202700 (mechanosensitive ion channel). Genetic components of salinity stress identified in this study may be explored to develop new rootstocks suitable for salinity-affected regions.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Welle PD, Mauter MS. High-resolution model for estimating the economic and policy implications of agricultural soil salinization in California. Environ. Res. Lett. 2017;12:094010. doi: 10.1088/1748-9326/aa848e. - DOI
-
- Butcher K, Wick A, Desutter T, Chatterjee A, Harmon J. Soil salinity: A threat to global food security. Agron. J. 2016;108:2189–2200. doi: 10.2134/agronj2016.06.0368. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

