Construction and validation of a novel gene signature for predicting the prognosis of osteosarcoma
- PMID: 35075228
- PMCID: PMC8786962
- DOI: 10.1038/s41598-022-05341-5
Construction and validation of a novel gene signature for predicting the prognosis of osteosarcoma
Abstract
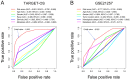
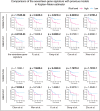
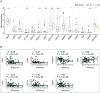
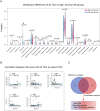
Osteosarcoma (OS) is the most common type of primary malignant bone tumor. The high-throughput sequencing technology has shown potential abilities to illuminate the pathogenic genes in OS. This study was designed to find a powerful gene signature that can predict clinical outcomes. We selected OS cases with gene expression and survival data in the TARGET-OS dataset and GSE21257 datasets as training cohort and validation cohort, respectively. The univariate Cox regression and Kaplan-Meier analysis were conducted to determine potential prognostic genes from the training cohort. These potential prognostic genes underwent a LASSO regression, which then generated a gene signature. The harvested signature's predictive ability was further examined by the Kaplan-Meier analysis, Cox analysis, and receiver operating characteristic (ROC curve). More importantly, we listed similar studies in the most recent year and compared theirs with ours. Finally, we performed functional annotation, immune relevant signature correlation identification, and immune infiltrating analysis to better study he functional mechanism of the signature and the immune cells' roles in the gene signature's prognosis ability. A seventeen-gene signature (UBE2L3, PLD3, SLC45A4, CLTC, CTNNBIP1, FBXL5, MKL2, SELPLG, C3orf14, WDR53, ZFP90, UHRF2, ARX, CORT, DDX26B, MYC, and SLC16A3) was generated from the LASSO regression. The signature was then confirmed having strong and stable prognostic capacity in all studied cohorts by several statistical methods. We revealed the superiority of our signature after comparing it to our predecessors, and the GO and KEGG annotations uncovered the specifically mechanism of action related to the gene signature. Six immune signatures, including PRF1, CD8A, HAVCR2, LAG3, CD274, and GZMA were identified associating with our signature. The immune-infiltrating analysis recognized the vital roles of T cells CD8 and Mast cells activated, which potentially support the seventeen-gene signature's prognosis ability. We identified a robust seventeen-gene signature that can accurately predict OS prognosis. We identified potential immunotherapy targets to the gene signature. The T cells CD8 and Mast cells activated were identified linked with the seventeen-gene signature predictive power.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
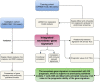
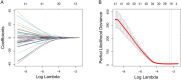
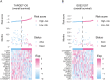
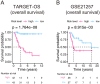


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

