Flanking sequences influence the activity of TET1 and TET2 methylcytosine dioxygenases and affect genomic 5hmC patterns
- PMID: 35075236
- PMCID: PMC8786823
- DOI: 10.1038/s42003-022-03033-4
Flanking sequences influence the activity of TET1 and TET2 methylcytosine dioxygenases and affect genomic 5hmC patterns
Abstract
TET dioxygenases convert 5-methylcytosine (5mC) preferentially in a CpG context into 5-hydroxymethylcytosine (5hmC) and higher oxidized forms, thereby initiating DNA demethylation, but details regarding the effects of the DNA sequences flanking the target 5mC site on TET activity are unknown. We investigated oxidation of libraries of DNA substrates containing one 5mC or 5hmC residue in randomized sequence context using single molecule readout of oxidation activity and sequence and show pronounced 20 and 70-fold flanking sequence effects on the catalytic activities of TET1 and TET2, respectively. Flanking sequence preferences were similar for TET1 and TET2 and also for 5mC and 5hmC substrates. Enhanced flanking sequence preferences were observed at non-CpG sites together with profound effects of flanking sequences on the specificity of TET2. TET flanking sequence preferences are reflected in genome-wide and local patterns of 5hmC and DNA demethylation in human and mouse cells indicating that they influence genomic DNA modification patterns in combination with locus specific targeting of TET enzymes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
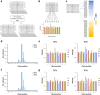


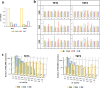


References
-
- Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. - PubMed
-
- Ambrosi C, Manzo M, Baubec T. Dynamics and context-dependent roles of DNA methylation. J. Mol. Biol. 2017;429:1459–1475. - PubMed
-
- Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem. Sci. 2014;39:310–318. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

