Lamina-specific immunohistochemical signatures in the olfactory bulb of healthy, Alzheimer's and Parkinson's disease patients
- PMID: 35075270
- PMCID: PMC8786934
- DOI: 10.1038/s42003-022-03032-5
Lamina-specific immunohistochemical signatures in the olfactory bulb of healthy, Alzheimer's and Parkinson's disease patients
Abstract
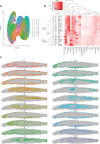
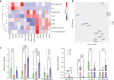
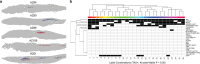
Traditional neuroanatomy immunohistology studies involve low-content analyses of a few antibodies of interest, typically applied and compared across sequential tissue sections. The efficiency, consistency, and ultimate insights of these studies can be substantially improved using high-plex immunofluorescence labelling on a single tissue section to allow direct comparison of many markers. Here we present an expanded and efficient multiplexed fluorescence-based immunohistochemistry (MP-IHC) approach that improves throughput with sequential labelling of up to 10 antibodies per cycle, with no limitation on the number of cycles, and maintains versatility and accessibility by using readily available commercial reagents and standard epifluorescence microscopy imaging. We demonstrate this approach by cumulatively screening up to 100 markers on formalin-fixed paraffin-embedded sections of human olfactory bulb sourced from neurologically normal (no significant pathology), Alzheimer's (AD), and Parkinson's disease (PD) patients. This brain region is involved early in the symptomology and pathophysiology of AD and PD. We also developed a spatial pixel bin analysis approach for unsupervised analysis of the high-content anatomical information from large tissue sections. Here, we present a comprehensive immunohistological characterisation of human olfactory bulb anatomy and a summary of differentially expressed biomarkers in AD and PD using the MP-IHC labelling and spatial protein analysis pipeline.
© 2022. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: A review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70:46–58. - PubMed
-
- Gut, G., Herrmann, M. D. & Pelkmans, L. Multiplexed protein maps link subcellular organization to cellular states. Science (New York, N.Y.) 361, 1–13 (2018). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

