This is a preprint.
SARS-CoV-2 Omicron spike mediated immune escape and tropism shift
- PMID: 35075452
- PMCID: PMC8786230
- DOI: 10.21203/rs.3.rs-1191837/v1
SARS-CoV-2 Omicron spike mediated immune escape and tropism shift
Update in
-
Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity.Nature. 2022 Mar;603(7902):706-714. doi: 10.1038/s41586-022-04474-x. Epub 2022 Feb 1. Nature. 2022. PMID: 35104837 Free PMC article.
Abstract
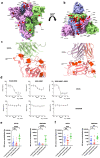
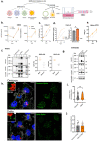
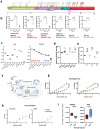
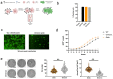
The SARS-CoV-2 Omicron BA.1 variant emerged in late 2021 and is characterised by multiple spike mutations across all spike domains. Here we show that Omicron BA.1 has higher affinity for ACE2 compared to Delta, and confers very significant evasion of therapeutic monoclonal and vaccine-elicited polyclonal neutralising antibodies after two doses. mRNA vaccination as a third vaccine dose rescues and broadens neutralisation. Importantly, antiviral drugs remdesevir and molnupiravir retain efficacy against Omicron BA.1. We found that in human nasal epithelial 3D cultures replication was similar for both Omicron and Delta. However, in lower airway organoids, Calu-3 lung cells and gut adenocarcinoma cell lines live Omicron virus demonstrated significantly lower replication in comparison to Delta. We noted that despite presence of mutations predicted to favour spike S1/S2 cleavage, the spike protein is less efficiently cleaved in live Omicron virions compared to Delta virions. We mapped the replication differences between the variants to entry efficiency using spike pseudotyped virus (PV) entry assays. The defect for Omicron PV in specific cell types correlated with higher cellular RNA expression of TMPRSS2, and accordingly knock down of TMPRSS2 impacted Delta entry to a greater extent as compared to Omicron. Furthermore, drug inhibitors targeting specific entry pathways demonstrated that the Omicron spike inefficiently utilises the cellular protease TMPRSS2 that mediates cell entry via plasma membrane fusion. Instead, we demonstrate that Omicron spike has greater dependency on cell entry via the endocytic pathway requiring the activity of endosomal cathepsins to cleave spike. Consistent with suboptimal S1/S2 cleavage and inability to utilise TMPRSS2, syncytium formation by the Omicron spike was dramatically impaired compared to the Delta spike. Overall, Omicron appears to have gained significant evasion from neutralising antibodies whilst maintaining sensitivity to antiviral drugs targeting the polymerase. Omicron has shifted cellular tropism away from TMPRSS2 expressing cells that are enriched in cells found in the lower respiratory and GI tracts, with implications for altered pathogenesis.
Figures
References
-
- Cele S. et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv, 2021.2012.2008.21267417, doi:10.1101/2021.12.08.21267417 (2021). - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

