Liquid biopsy reveals collateral tissue damage in cancer
- PMID: 35076021
- PMCID: PMC8855834
- DOI: 10.1172/jci.insight.153559
Liquid biopsy reveals collateral tissue damage in cancer
Abstract
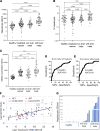
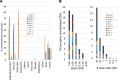
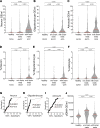
Cancer inflicts damage to surrounding normal tissues, which can culminate in fatal organ failure. Here, we demonstrate that cell death in organs affected by cancer can be detected by tissue-specific methylation patterns of circulating cell-free DNA (cfDNA). We detected elevated levels of hepatocyte-derived cfDNA in the plasma of patients with liver metastases originating from different primary tumors, compared with cancer patients without liver metastases. In addition, patients with localized pancreatic or colon cancer showed elevated hepatocyte cfDNA, suggesting liver damage inflicted by micrometastatic disease, by primary pancreatic tumor pressing the bile duct, or by a systemic response to the primary tumor. We also identified elevated neuron-, oligodendrocyte-, and astrocyte-derived cfDNA in a subpopulation of patients with brain metastases compared with cancer patients without brain metastasis. Cell type-specific cfDNA methylation markers enabled the identification of collateral tissue damage in cancer, revealing the presence of metastases in specific locations and potentially assisting in early cancer detection.
Keywords: Cell Biology; Epigenetics; Molecular diagnosis; Oncology.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

