Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement
- PMID: 35076256
- PMCID: PMC9427005
- DOI: 10.1126/science.abn8652
Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement
Abstract
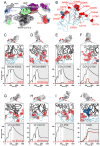
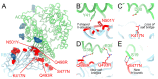
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant of concern evades antibody-mediated immunity that comes from vaccination or infection with earlier variants due to accumulation of numerous spike mutations. To understand the Omicron antigenic shift, we determined cryo-electron microscopy and x-ray crystal structures of the spike protein and the receptor-binding domain bound to the broadly neutralizing sarbecovirus monoclonal antibody (mAb) S309 (the parent mAb of sotrovimab) and to the human ACE2 receptor. We provide a blueprint for understanding the marked reduction of binding of other therapeutic mAbs that leads to dampened neutralizing activity. Remodeling of interactions between the Omicron receptor-binding domain and human ACE2 likely explains the enhanced affinity for the host receptor relative to the ancestral virus.
Figures


References
-
- Viana R., Moyo S., Amoako D. G., Tegally H., Scheepers C., Althaus C. L., Anyaneji U. J., Bester P. A., Boni M. F., Chand M., Choga W. T., Colquhoun R., Davids M., Deforche K., Doolabh D., du Plessis L., Engelbrecht S., Everatt J., Giandhari J., Giovanetti M., Hardie D., Hill V., Hsiao N.-Y., Iranzadeh A., Ismail A., Joseph C., Joseph R., Koopile L., Kosakovsky Pond S. L., Kraemer M. U. G., Kuate-Lere L., Laguda-Akingba O., Lesetedi-Mafoko O., Lessells R. J., Lockman S., Lucaci A. G., Maharaj A., Mahlangu B., Maponga T., Mahlakwane K., Makatini Z., Marais G., Maruapula D., Masupu K., Matshaba M., Mayaphi S., Mbhele N., Mbulawa M. B., Mendes A., Mlisana K., Mnguni A., Mohale T., Moir M., Moruisi K., Mosepele M., Motsatsi G., Motswaledi M. S., Mphoyakgosi T., Msomi N., Mwangi P. N., Naidoo Y., Ntuli N., Nyaga M., Olubayo L., Pillay S., Radibe B., Ramphal Y., Ramphal U., San J. E., Scott L., Shapiro R., Singh L., Smith-Lawrence P., Stevens W., Strydom A., Subramoney K., Tebeila N., Tshiabuila D., Tsui J., van Wyk S., Weaver S., Wibmer C. K., Wilkinson E., Wolter N., Zarebski A. E., Zuze B., Goedhals D., Preiser W., Treurnicht F., Venter M., Williamson C., Pybus O. G., Bhiman J., Glass A., Martin D. P., Rambaut A., Gaseitsiwe S., von Gottberg A., de Oliveira T., Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 10.1038/s41586-022-04411-y (2022). 10.1038/s41586-022-04411-y - DOI - PMC - PubMed
-
- McCallum M., Walls A. C., Sprouse K. R., Bowen J. E., Rosen L. E., Dang H. V., De Marco A., Franko N., Tilles S. W., Logue J., Miranda M. C., Ahlrichs M., Carter L., Snell G., Pizzuto M. S., Chu H. Y., Van Voorhis W. C., Corti D., Veesler D., Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 374, 1621–1626 (2021). 10.1126/science.abl8506 - DOI - PubMed
-
- McCallum M., Bassi J., De Marco A., Chen A., Walls A. C., Di Iulio J., Tortorici M. A., Navarro M.-J., Silacci-Fregni C., Saliba C., Sprouse K. R., Agostini M., Pinto D., Culap K., Bianchi S., Jaconi S., Cameroni E., Bowen J. E., Tilles S. W., Pizzuto M. S., Guastalla S. B., Bona G., Pellanda A. F., Garzoni C., Van Voorhis W. C., Rosen L. E., Snell G., Telenti A., Virgin H. W., Piccoli L., Corti D., Veesler D., SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 373, 648–654 (2021). 10.1126/science.abi7994 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

