Transcriptional Regulation and Implications for Controlling Hox Gene Expression
- PMID: 35076545
- PMCID: PMC8788451
- DOI: 10.3390/jdb10010004
Transcriptional Regulation and Implications for Controlling Hox Gene Expression
Abstract
Hox genes play key roles in axial patterning and regulating the regional identity of cells and tissues in a wide variety of animals from invertebrates to vertebrates. Nested domains of Hox expression generate a combinatorial code that provides a molecular framework for specifying the properties of tissues along the A-P axis. Hence, it is important to understand the regulatory mechanisms that coordinately control the precise patterns of the transcription of clustered Hox genes required for their roles in development. New insights are emerging about the dynamics and molecular mechanisms governing transcriptional regulation, and there is interest in understanding how these may play a role in contributing to the regulation of the expression of the clustered Hox genes. In this review, we summarize some of the recent findings, ideas and emerging mechanisms underlying the regulation of transcription in general and consider how they may be relevant to understanding the transcriptional regulation of Hox genes.
Keywords: Hox genes; coordinate regulation; enhancers; gene regulation; nascent transcripts; transcription factors; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

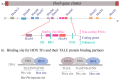
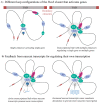
References
-
- Darras S., Fritzenwanker J.H., Uhlinger K.R., Farrelly E., Pani A.M., Hurley I.A., Norris R.P., Osovitz M., Terasaki M., Wu M., et al. Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol. 2018;16:e2003698. doi: 10.1371/journal.pbio.2003698. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

