Optimized Protocols for the Propagation and Quantification of Infectious Murine Hepatitis Virus (MHV-A59) Using NCTC Clone 1469 and 929 Cells
- PMID: 35076547
- PMCID: PMC8788426
- DOI: 10.3390/mps5010005
Optimized Protocols for the Propagation and Quantification of Infectious Murine Hepatitis Virus (MHV-A59) Using NCTC Clone 1469 and 929 Cells
Abstract
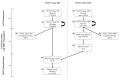
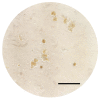
Murine hepatitis virus (MHV) is a non-human pathogen betacoronavirus that is evolutionarily and structurally related to the human pathogenic viruses SARS-CoV, MERS-CoV, and SARS-CoV-2. However, unlike the human SARS and MERS viruses, MHV requires a biosafety level 2 laboratory for propagating and safe handling, making it a potentially suitable surrogate virus. Despite this utility, few papers discussed the propagation and quantification of MHV using cell lines readily available in biorepositories making their implementations not easily reproducible. This article provides protocols for propagating and quantifying MHV-A59 using the recommended NCTC clone 1469 and clone 929 cell lines from American Type Culture Collection (ATCC). More specifically, the methods detail reviving cells, routine cell passaging, preparing freeze stocks, infection of NCTC clone 1469 with MHV and subsequent harvesting, and plaque assay quantification of MHV using NCTC clone 929 cells. Using these protocols, a BSL-2 laboratory equipped for cell culture work would generate at least 6.0 log plaque-forming units (PFU) per mL of MHV lysate and provide an optimized overlay assay using either methylcellulose or agarose as overlays for the titration of infectious virus particles. The protocols described here are intended to be utilized for persistence and inactivation studies of coronaviruses.
Keywords: CCL-1; CCL-9.1; COVID-19; MHV; SARS; cell passage; coronavirus; plaque assay; surrogate.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

