Antioxidant Response Activating nanoParticles (ARAPas) localize to atherosclerotic plaque and locally activate the Nrf2 pathway
- PMID: 35076645
- PMCID: PMC9181183
- DOI: 10.1039/d1bm01421h
Antioxidant Response Activating nanoParticles (ARAPas) localize to atherosclerotic plaque and locally activate the Nrf2 pathway
Abstract
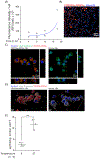
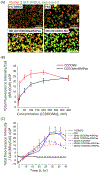
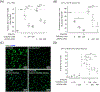
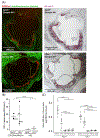
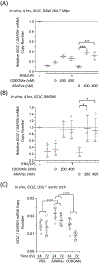
Atherosclerotic disease is the leading cause of death world-wide with few novel therapies available despite the ongoing health burden. Redox dysfunction is a well-established driver of atherosclerotic progression; however, the clinical translation of redox-based therapies is lacking. One of the challenges facing redox-based therapies is their targeted delivery to cellular domains of redox dysregulation. In the current study, we sought to develop Antioxidant Response Activating nanoParticles (ARAPas), encapsulating redox-based interventions, that exploit macrophage biology and the dysfunctional endothelium in order to selectively accumulate in atherosclerotic plaque. We employed flash nanoprecipitation (FNP) to synthesize bio-compatible polymeric nanoparticles encapsulating the hydrophobic Nrf2 activator drug, CDDO-Methyl (CDDOMe-ARAPas). Nuclear factor erythroid 2-related factor 2 (Nrf2)-activators are a promising class of redox-active drug molecules whereby activation of Nrf2 results in the expression of several antioxidant and cyto-protective enzymes that can be athero-protective. In this study, we characterize the physicochemical properties of CDDOMe-ARAPas as well as confirm their in vitro internalization by murine macrophages. Drug release of CDDOMe was determined by Nrf2-driven GFP fluorescence. Moreover, we show that these CDDOMe-ARAPas exert anti-inflammatory effects in classically activated macrophages. Finally, we show that CDDOMe-ARAPas selectively accumulate in atherosclerotic plaque of two widely-used murine models of atherosclerosis: ApoE-/- and LDLr-/- mice, and are capable of increasing gene expression of Nrf2-transcriptional targets in the atherosclerotic aortic arch. Future work will assess the therapeutic efficacy of intra-plaque Nrf2 activation with CDDOMe-ARAPas to inhibit atherosclerotic plaque progression. Overall, our present studies underline that targeting of atherosclerotic plaque is an effective means to enhance delivery of redox-based interventions.
Conflict of interest statement
Conflicts of interest
There are no conflicts to declare.
Figures




Similar articles
-
Alpinetin inhibits macrophage infiltration and atherosclerosis by improving the thiol redox state: Requirement of GSk3β/Fyn-dependent Nrf2 activation.FASEB J. 2022 Apr;36(4):e22261. doi: 10.1096/fj.202101567R. FASEB J. 2022. PMID: 35332570
-
Nuclear factor E2-related factor 2 deficiency impairs atherosclerotic lesion development but promotes features of plaque instability in hypercholesterolaemic mice.Cardiovasc Res. 2019 Jan 1;115(1):243-254. doi: 10.1093/cvr/cvy143. Cardiovasc Res. 2019. PMID: 29917052
-
Delivery of Cinnamic Aldehyde Antioxidant Response Activating nanoParticles (ARAPas) for Vascular Applications.Antioxidants (Basel). 2021 Apr 29;10(5):709. doi: 10.3390/antiox10050709. Antioxidants (Basel). 2021. PMID: 33946889 Free PMC article.
-
Combating oxidative stress in diabetic complications with Nrf2 activators: how much is too much?Redox Rep. 2014 May;19(3):107-17. doi: 10.1179/1351000214Y.0000000087. Epub 2014 Feb 21. Redox Rep. 2014. PMID: 24559141 Free PMC article. Review.
-
Counteracting role of nuclear factor erythroid 2-related factor 2 pathway in Alzheimer's disease.Biomed Pharmacother. 2020 Sep;129:110373. doi: 10.1016/j.biopha.2020.110373. Epub 2020 Jun 27. Biomed Pharmacother. 2020. PMID: 32603894 Review.
Cited by
-
Design of Modular, 3D-Printed Millifluidic Mixers to Enable Sequential NanoPrecipitation (SNaP) for the Tunable Synthesis of Drug-Loaded Nanoparticles and Microparticles.Adv Mater Technol. 2025 Jan 8;10(1):2400583. doi: 10.1002/admt.202570002. Epub 2024 Aug 3. Adv Mater Technol. 2025. PMID: 40814357 Free PMC article.
-
The Use of Acute Immunosuppressive Therapy to Improve Antibiotic Efficacy against Intracellular Staphylococcus aureus.Microbiol Spectr. 2022 Jun 29;10(3):e0085822. doi: 10.1128/spectrum.00858-22. Epub 2022 May 16. Microbiol Spectr. 2022. PMID: 35575507 Free PMC article.
-
Multiple myeloma inhibitory effects of natural compounds: enhancement through nanoparticle carriers.Front Pharmacol. 2025 Jun 17;16:1589090. doi: 10.3389/fphar.2025.1589090. eCollection 2025. Front Pharmacol. 2025. PMID: 40599807 Free PMC article. Review.
-
Role of NRF2 in Pathogenesis of Alzheimer's Disease.Antioxidants (Basel). 2024 Dec 13;13(12):1529. doi: 10.3390/antiox13121529. Antioxidants (Basel). 2024. PMID: 39765857 Free PMC article. Review.
-
Advances in Nrf2 Signaling Pathway by Targeted Nanostructured-Based Drug Delivery Systems.Biomedicines. 2024 Feb 9;12(2):403. doi: 10.3390/biomedicines12020403. Biomedicines. 2024. PMID: 38398005 Free PMC article. Review.
References
-
- W. H. Organization, 2017.
-
- Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jiménez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS and Muntner P, Circulation, 2017, 135, e146–e603. - PMC - PubMed
-
- Libby P, Buring JE, Badimon L, Hansson GK, Deanfield J, Bittencourt MS, Tokgözoğlu L and Lewis EF, Nat Rev Dis Primers, 2019, 5, 56. - PubMed
-
- Hansson GK and Hermansson A, Nat Immunol, 2011, 12, 204–212. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

