The estimates of effective population size based on linkage disequilibrium are virtually unaffected by natural selection
- PMID: 35077457
- PMCID: PMC8815936
- DOI: 10.1371/journal.pgen.1009764
The estimates of effective population size based on linkage disequilibrium are virtually unaffected by natural selection
Abstract
The effective population size (Ne) is a key parameter to quantify the magnitude of genetic drift and inbreeding, with important implications in human evolution. The increasing availability of high-density genetic markers allows the estimation of historical changes in Ne across time using measures of genome diversity or linkage disequilibrium between markers. Directional selection is expected to reduce diversity and Ne, and this reduction is modulated by the heterogeneity of the genome in terms of recombination rate. Here we investigate by computer simulations the consequences of selection (both positive and negative) and recombination rate heterogeneity in the estimation of historical Ne. We also investigate the relationship between diversity parameters and Ne across the different regions of the genome using human marker data. We show that the estimates of historical Ne obtained from linkage disequilibrium between markers (NeLD) are virtually unaffected by selection. In contrast, those estimates obtained by coalescence mutation-recombination-based methods can be strongly affected by it, which could have important consequences for the estimation of human demography. The simulation results are supported by the analysis of human data. The estimates of NeLD obtained for particular genomic regions do not correlate, or they do it very weakly, with recombination rate, nucleotide diversity, proportion of polymorphic sites, background selection statistic, minor allele frequency of SNPs, loss of function and missense variants and gene density. This suggests that NeLD measures mainly reflect demographic changes in population size across generations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
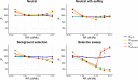



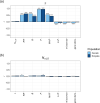
Similar articles
-
Estimation of linkage disequilibrium and effective population size in New Zealand sheep using three different methods to create genetic maps.BMC Genet. 2017 Jul 21;18(1):68. doi: 10.1186/s12863-017-0534-2. BMC Genet. 2017. PMID: 28732466 Free PMC article.
-
Genetic diversity of a New Zealand multi-breed sheep population and composite breeds' history revealed by a high-density SNP chip.BMC Genet. 2017 Mar 14;18(1):25. doi: 10.1186/s12863-017-0492-8. BMC Genet. 2017. PMID: 28288558 Free PMC article.
-
Making sense of genetic estimates of effective population size.Mol Ecol. 2016 Oct;25(19):4689-91. doi: 10.1111/mec.13814. Mol Ecol. 2016. PMID: 27671356
-
Prediction and estimation of effective population size.Heredity (Edinb). 2016 Oct;117(4):193-206. doi: 10.1038/hdy.2016.43. Epub 2016 Jun 29. Heredity (Edinb). 2016. PMID: 27353047 Free PMC article. Review.
-
Linkage disequilibrium: ancient history drives the new genetics.Hum Hered. 2005;59(2):118-24. doi: 10.1159/000085226. Epub 2005 Apr 18. Hum Hered. 2005. PMID: 15838181 Review.
Cited by
-
Genomic diversity of the locally developed Latvian Darkheaded sheep breed.Heliyon. 2024 May 16;10(10):e31455. doi: 10.1016/j.heliyon.2024.e31455. eCollection 2024 May 30. Heliyon. 2024. PMID: 38807890 Free PMC article.
-
Biases in ARG-Based Inference of Historical Population Size in Populations Experiencing Selection.Mol Biol Evol. 2024 Jul 3;41(7):msae118. doi: 10.1093/molbev/msae118. Mol Biol Evol. 2024. PMID: 38874402 Free PMC article.
-
Integrating evolutionary genomics of forest trees to inform future tree breeding amid rapid climate change.Plant Commun. 2024 Oct 14;5(10):101044. doi: 10.1016/j.xplc.2024.101044. Epub 2024 Aug 7. Plant Commun. 2024. PMID: 39095989 Free PMC article. Review.
-
Estimating the optimal number of samples to determine the effective population size in livestock.Front Genet. 2025 Jun 3;16:1588986. doi: 10.3389/fgene.2025.1588986. eCollection 2025. Front Genet. 2025. PMID: 40529807 Free PMC article.
-
Trustworthy causal biomarker discovery: a multiomics brain imaging genetics-based approach.Bioinformatics. 2025 Jul 1;41(Supplement_1):i227-i236. doi: 10.1093/bioinformatics/btaf257. Bioinformatics. 2025. PMID: 40662843 Free PMC article.
References
-
- Caballero A. Quantitative Genetics. Cambridge University Press; 2020.
-
- Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW. Estimation of census and effective population sizes: The increasing usefulness of DNA-based approaches. Conserv Genet. 2010; 11: 355–373.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

