AMPlify: attentive deep learning model for discovery of novel antimicrobial peptides effective against WHO priority pathogens
- PMID: 35078402
- PMCID: PMC8788131
- DOI: 10.1186/s12864-022-08310-4
AMPlify: attentive deep learning model for discovery of novel antimicrobial peptides effective against WHO priority pathogens
Abstract
Background: Antibiotic resistance is a growing global health concern prompting researchers to seek alternatives to conventional antibiotics. Antimicrobial peptides (AMPs) are attracting attention again as therapeutic agents with promising utility in this domain, and using in silico methods to discover novel AMPs is a strategy that is gaining interest. Such methods can sift through large volumes of candidate sequences and reduce lab screening costs.
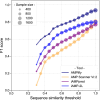
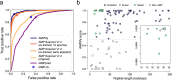
Results: Here we introduce AMPlify, an attentive deep learning model for AMP prediction, and demonstrate its utility in prioritizing peptide sequences derived from the Rana [Lithobates] catesbeiana (bullfrog) genome. We tested the bioactivity of our predicted peptides against a panel of bacterial species, including representatives from the World Health Organization's priority pathogens list. Four of our novel AMPs were active against multiple species of bacteria, including a multi-drug resistant isolate of carbapenemase-producing Escherichia coli.
Conclusions: We demonstrate the utility of deep learning based tools like AMPlify in our fight against antibiotic resistance. We expect such tools to play a significant role in discovering novel candidates of peptide-based alternatives to classical antibiotics.
Keywords: Antimicrobial peptide; Attention mechanism; Deep learning.
© 2022. The Author(s).
Conflict of interest statement
IB is a co-founder of and executive at Amphoraxe Life Sciences Inc.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

