The mixed blessing of AMPK signaling in Cancer treatments
- PMID: 35078427
- PMCID: PMC8786626
- DOI: 10.1186/s12885-022-09211-1
The mixed blessing of AMPK signaling in Cancer treatments
Abstract
Background: Nutrient acquisition and metabolism pathways are altered in cancer cells to meet bioenergetic and biosynthetic demands. A major regulator of cellular metabolism and energy homeostasis, in normal and cancer cells, is AMP-activated protein kinase (AMPK). AMPK influences cell growth via its modulation of the mechanistic target of Rapamycin (mTOR) pathway, specifically, by inhibiting mTOR complex mTORC1, which facilitates cell proliferation, and by activating mTORC2 and cell survival. Given its conflicting roles, the effects of AMPK activation in cancer can be counter intuitive. Prior to the establishment of cancer, AMPK acts as a tumor suppressor. However, following the onset of cancer, AMPK has been shown to either suppress or promote cancer, depending on cell type or state.
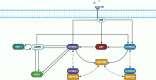
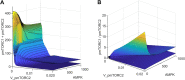
Methods: To unravel the controversial roles of AMPK in cancer, we developed a computational model to simulate the effects of pharmacological maneuvers that target key metabolic signalling nodes, with a specific focus on AMPK, mTORC, and their modulators. Specifically, we constructed an ordinary differential equation-based mechanistic model of AMPK-mTORC signaling, and parametrized the model based on existing experimental data.
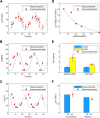
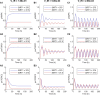
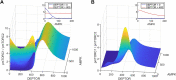
Results: Model simulations were conducted to yield the following predictions: (i) increasing AMPK activity has opposite effects on mTORC depending on the nutrient availability; (ii) indirect inhibition of AMPK activity through inhibition of sirtuin 1 (SIRT1) only has an effect on mTORC activity under conditions of low nutrient availability; (iii) the balance between cell proliferation and survival exhibits an intricate dependence on DEP domain-containing mTOR-interacting protein (DEPTOR) abundance and AMPK activity; (iv) simultaneous direct inhibition of mTORC2 and activation of AMPK is a potential strategy for suppressing both cell survival and proliferation.
Conclusions: Taken together, model simulations clarify the competing effects and the roles of key metabolic signalling pathways in tumorigenesis, which may yield insights on innovative therapeutic strategies.
Keywords: AMPK; Cancer; Dynamical system; Metabolism; mTORC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests
Figures


References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

