Extracellular vimentin is an attachment factor that facilitates SARS-CoV-2 entry into human endothelial cells
- PMID: 35078919
- PMCID: PMC8833221
- DOI: 10.1073/pnas.2113874119
Extracellular vimentin is an attachment factor that facilitates SARS-CoV-2 entry into human endothelial cells
Abstract
SARS-CoV-2 entry into host cells is a crucial step for virus tropism, transmission, and pathogenesis. Angiotensin-converting enzyme 2 (ACE2) has been identified as the primary entry receptor for SARS-CoV-2; however, the possible involvement of other cellular components in the viral entry has not yet been fully elucidated. Here we describe the identification of vimentin (VIM), an intermediate filament protein widely expressed in cells of mesenchymal origin, as an important attachment factor for SARS-CoV-2 on human endothelial cells. Using liquid chromatography-tandem mass spectrometry, we identified VIM as a protein that binds to the SARS-CoV-2 spike (S) protein. We showed that the S-protein receptor binding domain (RBD) is sufficient for S-protein interaction with VIM. Further analysis revealed that extracellular VIM binds to SARS-CoV-2 S-protein and facilitates SARS-CoV-2 infection, as determined by entry assays performed with pseudotyped viruses expressing S and with infectious SARS-CoV-2. Coexpression of VIM with ACE2 increased SARS-CoV-2 entry in HEK-293 cells, and shRNA-mediated knockdown of VIM significantly reduced SARS-CoV-2 infection of human endothelial cells. Moreover, incubation of A549 cells expressing ACE2 with purified VIM increased pseudotyped SARS-CoV-2-S entry. CR3022 antibody, which recognizes a distinct epitope on SARS-CoV-2-S-RBD without interfering with the binding of the spike with ACE2, inhibited the binding of VIM with CoV-2 S-RBD, and neutralized viral entry in human endothelial cells, suggesting a key role for VIM in SARS-CoV-2 infection of endothelial cells. This work provides insight into the pathogenesis of COVID-19 linked to the vascular system, with implications for the development of therapeutics and vaccines.
Keywords: ACE2; SARS-CoV-2; endothelial cells; vimentin; viral entry.
Copyright © 2022 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures




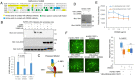
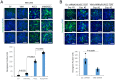
References
-
- Lan J., et al. , Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

