Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer
- PMID: 35078920
- PMCID: PMC8833191
- DOI: 10.1073/pnas.2121644119
Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer
Abstract
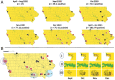
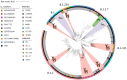
Many animal species are susceptible to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and could act as reservoirs; however, transmission in free-living animals has not been documented. White-tailed deer, the predominant cervid in North America, are susceptible to SARS-CoV-2 infection, and experimentally infected fawns can transmit the virus. To test the hypothesis that SARS-CoV-2 is circulating in deer, 283 retropharyngeal lymph node (RPLN) samples collected from 151 free-living and 132 captive deer in Iowa from April 2020 through January of 2021 were assayed for the presence of SARS-CoV-2 RNA. Ninety-four of the 283 (33.2%) deer samples were positive for SARS-CoV-2 RNA as assessed by RT-PCR. Notably, following the November 2020 peak of human cases in Iowa, and coinciding with the onset of winter and the peak deer hunting season, SARS-CoV-2 RNA was detected in 80 of 97 (82.5%) RPLN samples collected over a 7-wk period. Whole genome sequencing of all 94 positive RPLN samples identified 12 SARS-CoV-2 lineages, with B.1.2 (n = 51; 54.5%) and B.1.311 (n = 19; 20%) accounting for ∼75% of all samples. The geographic distribution and nesting of clusters of deer and human lineages strongly suggest multiple human-to-deer transmission events followed by subsequent deer-to-deer spread. These discoveries have important implications for the long-term persistence of the SARS-CoV-2 pandemic. Our findings highlight an urgent need for a robust and proactive "One Health" approach to obtain enhanced understanding of the ecology, molecular evolution, and dissemination of SARS-CoV-2.
Keywords: One Health; SARS-CoV-2; animal reservoir; deer; spillover.
Copyright © 2022 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

Update of
-
Detection of SARS-CoV-2 Omicron variant (B.1.1.529) infection of white-tailed deer.bioRxiv [Preprint]. 2022 Feb 7:2022.02.04.479189. doi: 10.1101/2022.02.04.479189. bioRxiv. 2022. Update in: Proc Natl Acad Sci U S A. 2022 Feb 8;119(6):e2121644119. doi: 10.1073/pnas.2121644119. PMID: 35169802 Free PMC article. Updated. Preprint.
References
-
- Munster V. J., Koopmans M., van Doremalen N., van Riel D., de Wit E., A novel coronavirus emerging in China - Key questions for impact assessment. N. Engl. J. Med. 382, 692–694 (2020). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

