Whole-genome sequencing reveals the evolutionary trajectory of HBV-related hepatocellular carcinoma early recurrence
- PMID: 35078970
- PMCID: PMC8789859
- DOI: 10.1038/s41392-021-00838-3
Whole-genome sequencing reveals the evolutionary trajectory of HBV-related hepatocellular carcinoma early recurrence
Abstract
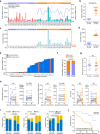
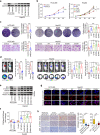
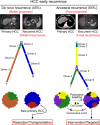
Patients with hepatocellular carcinoma (HCC) have poor long-term survival following curative resection because of the high rate of tumor early recurrence. Little is known about the trajectory of genomic evolution from primary to early-recurrent HCC. In this study, we performed whole-genome sequencing (WGS) on 40 pairs of primary and early-recurrent hepatitis B virus (HBV)-related HCC tumors from patients who received curative resection, and from four patients whose primary and recurrent tumor were extensively sampled. We identified two recurrence patterns: de novo recurrence (18/40), which developed genetically independently of the primary tumor and carried different HCC drivers, and ancestral recurrence (22/40), which was clonally related to the primary tumor and progressed more rapidly than de novo recurrence. We found that the recurrence location was predictive of the recurrence pattern: distant recurrence tended to display the de novo pattern, whereas local recurrence tended to display the ancestral pattern. We then uncovered the evolutionary trajectories based on the subclonal architecture, driver-gene mutations, and mutational processes observed in the primary and recurrent tumors. Multi-region WGS demonstrated spatiotemporal heterogeneity and polyclonal, monophyletic dissemination in HCC ancestral recurrence. In addition, we identified recurrence-specific mutations and copy-number gains in BCL9, leading to WNT/β-catenin signaling activation and an immune-excluded tumor microenvironment, which suggests that BCL9 might serve as a new therapeutic target for recurrent HCC. Collectively, our results allow us to view with unprecedented clarity the genomic evolution during HBV-related HCC early recurrence, providing an important molecular foundation for enhanced understanding of HCC with implications for personalized therapy to improve patient survival.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: Cancer J. Clinicians. 2018;68:7–30. - PubMed
-
- Ahn SM, et al. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–1982. - PubMed
-
- Fujimoto A, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016;48:500–509. - PubMed
-
- Totoki Y, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014;46:1267–1273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

