Coding and regulatory variants are associated with serum protein levels and disease
- PMID: 35079000
- PMCID: PMC8789809
- DOI: 10.1038/s41467-022-28081-6
Coding and regulatory variants are associated with serum protein levels and disease
Abstract
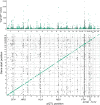
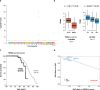
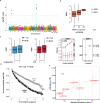
Circulating proteins can be used to diagnose and predict disease-related outcomes. A deep serum proteome survey recently revealed close associations between serum protein networks and common disease. In the current study, 54,469 low-frequency and common exome-array variants were compared to 4782 protein measurements in the serum of 5343 individuals from the AGES Reykjavik cohort. This analysis identifies a large number of serum proteins with genetic signatures overlapping those of many diseases. More specifically, using a study-wide significance threshold, we find that 2021 independent exome array variants are associated with serum levels of 1942 proteins. These variants reside in genetic loci shared by hundreds of complex disease traits, highlighting serum proteins' emerging role as biomarkers and potential causative agents of a wide range of diseases.
© 2022. The Author(s).
Conflict of interest statement
The study was supported by the Novartis Institute for Biomedical Research, and protein measurements for the AGES-RS cohort were performed at SomaLogic. J.R.L. and L.L.J. are employees and stockholders of Novartis. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

