Aptamer based point of care diagnostic for the detection of food allergens
- PMID: 35079047
- PMCID: PMC8789827
- DOI: 10.1038/s41598-022-05265-0
Aptamer based point of care diagnostic for the detection of food allergens
Abstract
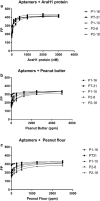
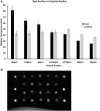
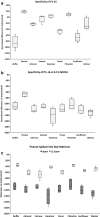
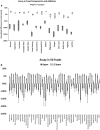
Aptamers, due to their small size, strong target affinity, and ease of chemical modification, are ideally suited for molecular detection technologies. Here, we describe successful use of aptamer technology in a consumer device for the detection of peanut antigen in food. The novel aptamer-based protein detection method is robust across a wide variety of food matrices and sensitive to peanut protein at concentrations as low as 12.5 ppm (37.5 µg peanut protein in the sample). Integration of the assay into a sensitive, stable, and consumer friendly portable device will empower users to easily and quickly assess the presence of peanut allergens in foods before eating. With many food reactions occurring outside the home, the type of technology described here has significant potential to improve lives for children and families.
© 2022. The Author(s).
Conflict of interest statement
S.S., V.V., V.C., O.A., L.Y., and A.G. were employed by DOTS Technology Corp during the work. A.G. is a co-founder and member of the board of directors. W.S., J.S., D.F., and H.S. are members of the scientific advisory board.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

