Prediction of allosteric sites and signaling: Insights from benchmarking datasets
- PMID: 35079717
- PMCID: PMC8767309
- DOI: 10.1016/j.patter.2021.100408
Prediction of allosteric sites and signaling: Insights from benchmarking datasets
Abstract
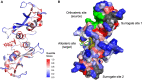
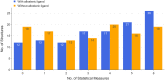
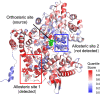
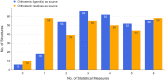
Allostery is a pervasive mechanism that regulates protein activity through ligand binding at a site different from the orthosteric site. The universality of allosteric regulation complemented by the benefits of highly specific and potentially non-toxic allosteric drugs makes uncovering allosteric sites invaluable. However, there are few computational methods to effectively predict them. Bond-to-bond propensity analysis has successfully predicted allosteric sites in 19 of 20 cases using an energy-weighted atomistic graph. We here extended the analysis onto 432 structures of 146 proteins from two benchmarking datasets for allosteric proteins: ASBench and CASBench. We further introduced two statistical measures to account for the cumulative effect of high-propensity residues and the crucial residues in a given site. The allosteric site is recovered for 127 of 146 proteins (407 of 432 structures) knowing only the orthosteric sites or ligands. The quantitative analysis using a range of statistical measures enables better characterization of potential allosteric sites and mechanisms involved.
Keywords: allosteric site detection; benchmarking; graph theory measures.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
AI in drug discovery: Applications, opportunities, and challenges.Patterns (N Y). 2022 Jun 10;3(6):100529. doi: 10.1016/j.patter.2022.100529. eCollection 2022 Jun 10. Patterns (N Y). 2022. PMID: 35755871 Free PMC article. No abstract available.
References
-
- Casem M.L. Academic Press; 2016. Chapter 3 - Proteins. Case Studies in Cell Biology; pp. 23–71. - DOI
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

