Disruption of pancreatic stellate cell myofibroblast phenotype promotes pancreatic tumor invasion
- PMID: 35081338
- PMCID: PMC8810397
- DOI: 10.1016/j.celrep.2021.110227
Disruption of pancreatic stellate cell myofibroblast phenotype promotes pancreatic tumor invasion
Abstract
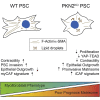
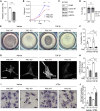
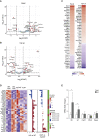
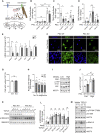
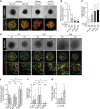
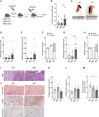
In pancreatic ductal adenocarcinoma (PDAC), differentiation of pancreatic stellate cells (PSCs) into myofibroblast-like cancer-associated fibroblasts (CAFs) can both promote and suppress tumor progression. Here, we show that the Rho effector protein kinase N2 (PKN2) is critical for PSC myofibroblast differentiation. Loss of PKN2 is associated with reduced PSC proliferation, contractility, and alpha-smooth muscle actin (α-SMA) stress fibers. In spheroid co-cultures with PDAC cells, loss of PKN2 prevents PSC invasion but, counter-intuitively, promotes invasive cancer cell outgrowth. PKN2 deletion induces a myofibroblast to inflammatory CAF switch in the PSC matrisome signature both in vitro and in vivo. Further, deletion of PKN2 in the pancreatic stroma induces more locally invasive, orthotopic pancreatic tumors. Finally, we demonstrate that a PKN2KO matrisome signature predicts poor outcome in pancreatic and other solid human cancers. Our data indicate that suppressing PSC myofibroblast function can limit important stromal tumor-suppressive mechanisms, while promoting a switch to a cancer-supporting CAF phenotype.
Keywords: CAF; PKN2; Rho GTPases; cancer-associated fibroblasts; matrisome; pancreatic cancer; protein kinase N2; tumour microenvironment.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Bachem M.G., Schneider E., Gross H., Weidenbach H., Schmid R.M., Menke A., Siech M., Beger H., Grunert A., Adler G. Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology. 1998;115:421–432. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

