Identification of a novel HOOK3-FGFR1 fusion gene involved in activation of the NF-kappaB pathway
- PMID: 35081975
- PMCID: PMC8793161
- DOI: 10.1186/s12935-022-02451-y
Identification of a novel HOOK3-FGFR1 fusion gene involved in activation of the NF-kappaB pathway
Abstract
Background: Rearrangements involving the fibroblast growth factor receptor 1 (FGFR1) gene result in 8p11 myeloproliferative syndrome (EMS), which is a rare and aggressive hematological malignancy that is often initially diagnosed as myelodysplastic syndrome (MDS). Clinical outcomes are typically poor due to relative resistance to tyrosine kinase inhibitors (TKIs) and rapid transformation to acute leukemia. Deciphering the transcriptomic signature of FGFR1 fusions may open new treatment strategies for FGFR1 rearrangement patients.
Methods: DNA sequencing (DNA-seq) was performed for 20 MDS patients and whole exome sequencing (WES) was performed for one HOOK3-FGFR1 fusion positive patient. RNA sequencing (RNA-seq) was performed for 20 MDS patients and 8 healthy donors. Fusion genes were detected using the STAR-Fusion tool. Fluorescence in situ hybridization (FISH), quantitative real-time PCR (qRT-PCR), and Sanger sequencing were used to confirm the HOOK3-FGFR1 fusion gene. The phosphorylation antibody array was performed to validate the activation of nuclear factor-kappaB (NF-kappaB) signaling.
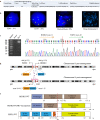
Results: We identified frequently recurrent mutations of ASXL1 and U2AF1 in the MDS cohort, which is consistent with previous reports. We also identified a novel in-frame HOOK3-FGFR1 fusion gene in one MDS case with abnormal monoclonal B-cell lymphocytosis and ring chromosome 8. FISH analysis detected the FGFR1 break-apart signal in myeloid blasts only. qRT-PCR and Sanger sequencing confirmed the HOOK3-FGFR1 fusion transcript with breakpoints located at the 11th exon of HOOK3 and 10th exon of FGFR1, and Western blot detected the chimeric HOOK3-FGFR1 fusion protein that is presumed to retain the entire tyrosine kinase domain of FGFR1. The transcriptional feature of HOOK3-FGFR1 fusion was characterized by the significant enrichment of the NF-kappaB pathway by comparing the expression profiling of FGFR1 fusion positive MDS with 8 healthy donors and FGFR1 fusion negative MDS patients. Further validation by phosphorylation antibody array also showed NF-kappaB activation, as evidenced by increased phosphorylation of p65 (Ser 536) and of IKBalpha (Ser 32).
Conclusions: The HOOK3-FGFR1 fusion gene may contribute to the pathogenesis of MDS and activate the NF-kappaB pathway. These findings highlight a potential novel approach for combination therapy for FGFR1 rearrangement patients.
Keywords: 8p11 myeloproliferative syndrome; Expression signature; HOOK3-FGFR1; NF-kappaB signaling pathway; RNA sequencing.
© 2022. The Author(s).
Conflict of interest statement
All authors have approved this submission and declare no competing interests.
Figures



References
-
- Jackson CC, Medeiros LJ, Miranda RN. 8p11 myeloproliferative syndrome: a review. Hum Pathol. 2010;41(4):461–76. - PubMed
-
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. - PubMed
-
- Umino K, Fujiwara SI, Ikeda T, Toda Y, Ito S, Mashima K, Minakata D, Nakano H, Yamasaki R, Kawasaki Y, et al. Clinical outcomes of myeloid/lymphoid neoplasms with fibroblast growth factor receptor-1 (FGFR1) rearrangement. Hematology. 2018;23(8):470–477. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

