In Silico Prediction and Insights Into the Structural Basis of Drug Induced Nephrotoxicity
- PMID: 35082675
- PMCID: PMC8785686
- DOI: 10.3389/fphar.2021.793332
In Silico Prediction and Insights Into the Structural Basis of Drug Induced Nephrotoxicity
Abstract
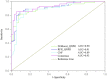
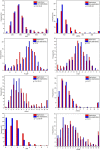
Drug induced nephrotoxicity is a major clinical challenge, and it is always associated with higher costs for the pharmaceutical industry and due to detection during the late stages of drug development. It is desirable for improving the health outcomes for patients to distinguish nephrotoxic structures at an early stage of drug development. In this study, we focused on in silico prediction and insights into the structural basis of drug induced nephrotoxicity, based on reliable data on human nephrotoxicity. We collected 565 diverse chemical structures, including 287 nephrotoxic drugs on humans in the real world, and 278 non-nephrotoxic approved drugs. Several different machine learning and deep learning algorithms were employed for in silico model building. Then, a consensus model was developed based on three best individual models (RFR_QNPR, XGBOOST_QNPR, and CNF). The consensus model performed much better than individual models on internal validation and it achieved prediction accuracy of 86.24% external validation. The results of analysis of molecular properties differences between nephrotoxic and non-nephrotoxic structures indicated that several key molecular properties differ significantly, including molecular weight (MW), molecular polar surface area (MPSA), AlogP, number of hydrogen bond acceptors (nHBA), molecular solubility (LogS), the number of rotatable bonds (nRotB), and the number of aromatic rings (nAR). These molecular properties may be able to play an important part in the identification of nephrotoxic chemicals. Finally, 87 structural alerts for chemical nephrotoxicity were mined with f-score and positive rate analysis of substructures from Klekota-Roth fingerprint (KRFP). These structural alerts can well identify nephrotoxic drug structures in the data set. The in silico models and the structural alerts could be freely accessed via https://ochem.eu/article/140251 and http://www.sapredictor.cn, respectively. We hope the results should provide useful tools for early nephrotoxicity estimation in drug development.
Keywords: consensus model; drug induced nephrotoxicity; in silico prediction; structural alert; web-server.
Copyright © 2022 Shi, Hua, Wang, Zhang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Breiman L. (2001). Random Forests. Machine Learn. 45 (1), 5–32. 10.1023/a:1010933404324 - DOI
-
- Capela F., Nouchi V., Van Deursen R., Tetko I. V., Godin G. (2019). Multitask Learning on Graph Neural Networks Applied to Molecular Property Predictions. Available at: https://ui.adsabs.harvard.edu/abs/2019arXiv191013124C (Accessed October 01, 2019).
LinkOut - more resources
Full Text Sources

