Persistence of SARS CoV-2 S1 Protein in CD16+ Monocytes in Post-Acute Sequelae of COVID-19 (PASC) up to 15 Months Post-Infection
- PMID: 35082777
- PMCID: PMC8784688
- DOI: 10.3389/fimmu.2021.746021
Persistence of SARS CoV-2 S1 Protein in CD16+ Monocytes in Post-Acute Sequelae of COVID-19 (PASC) up to 15 Months Post-Infection
Abstract
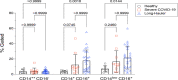
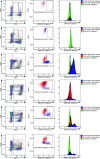
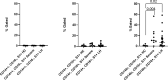
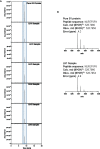
The recent COVID-19 pandemic is a treatment challenge in the acute infection stage but the recognition of chronic COVID-19 symptoms termed post-acute sequelae SARS-CoV-2 infection (PASC) may affect up to 30% of all infected individuals. The underlying mechanism and source of this distinct immunologic condition three months or more after initial infection remains elusive. Here, we investigated the presence of SARS-CoV-2 S1 protein in 46 individuals. We analyzed T-cell, B-cell, and monocytic subsets in both severe COVID-19 patients and in patients with post-acute sequelae of COVID-19 (PASC). The levels of both intermediate (CD14+, CD16+) and non-classical monocyte (CD14Lo, CD16+) were significantly elevated in PASC patients up to 15 months post-acute infection compared to healthy controls (P=0.002 and P=0.01, respectively). A statistically significant number of non-classical monocytes contained SARS-CoV-2 S1 protein in both severe (P=0.004) and PASC patients (P=0.02) out to 15 months post-infection. Non-classical monocytes were sorted from PASC patients using flow cytometric sorting and the SARS-CoV-2 S1 protein was confirmed by mass spectrometry. Cells from 4 out of 11 severe COVID-19 patients and 1 out of 26 PASC patients contained ddPCR+ peripheral blood mononuclear cells, however, only fragmented SARS-CoV-2 RNA was found in PASC patients. No full length sequences were identified, and no sequences that could account for the observed S1 protein were identified in any patient. That non-classical monocytes may be a source of inflammation in PASC warrants further study.
Keywords: CCR5; COVID-19; PASC; SARS CoV-2 S1 protein; fractalkine; non-classical monocytes.
Copyright © 2022 Patterson, Francisco, Yogendra, Long, Pise, Rodrigues, Hall, Herrera, Parikh, Guevara-Coto, Triche, Scott, Hekmati, Maglinte, Chang, Mora-Rodríguez and Mora.
Conflict of interest statement
BP, AP, HR, EL, and EF are employees of IncellDx, Inc. TT, PS, SH, and DM are employees of Avrok Laboratories, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- (2021). Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-care/post-covid-c....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

