Synthetic Biology Approaches to Hydrocarbon Biosensors: A Review
- PMID: 35083206
- PMCID: PMC8784404
- DOI: 10.3389/fbioe.2021.804234
Synthetic Biology Approaches to Hydrocarbon Biosensors: A Review
Abstract
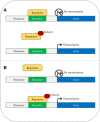
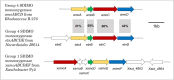
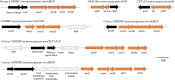
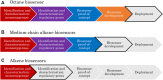
Monooxygenases are a class of enzymes that facilitate the bacterial degradation of alkanes and alkenes. The regulatory components associated with monooxygenases are nature's own hydrocarbon sensors, and once functionally characterised, these components can be used to create rapid, inexpensive and sensitive biosensors for use in applications such as bioremediation and metabolic engineering. Many bacterial monooxygenases have been identified, yet the regulation of only a few of these have been investigated in detail. A wealth of genetic and functional diversity of regulatory enzymes and promoter elements still remains unexplored and unexploited, both in published genome sequences and in yet-to-be-cultured bacteria. In this review we examine in detail the current state of research on monooxygenase gene regulation, and on the development of transcription-factor-based microbial biosensors for detection of alkanes and alkenes. A new framework for the systematic characterisation of the underlying genetic components and for further development of biosensors is presented, and we identify focus areas that should be targeted to enable progression of more biosensor candidates to commercialisation and deployment in industry and in the environment.
Keywords: alkane; alkene; bacteria; biosensor; hydrocarbon; monooxygenase; regulation; transcription factor.
Copyright © 2022 Moratti, Scott and Coleman.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

