Simulated Microgravity Potentiates Hematopoietic Differentiation of Human Pluripotent Stem Cells and Supports Formation of 3D Hematopoietic Cluster
- PMID: 35083220
- PMCID: PMC8784808
- DOI: 10.3389/fcell.2021.797060
Simulated Microgravity Potentiates Hematopoietic Differentiation of Human Pluripotent Stem Cells and Supports Formation of 3D Hematopoietic Cluster
Abstract
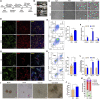
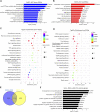
Microgravity has been shown to induces many changes in proliferation, differentiation and growth behavior of stem cells. Little is known about the effect of microgravity on hematopoietic differentiation of pluripotent stem cells (PSCs). In this study, we used the random position machine (RPM) to investigate whether simulated microgravity (SMG) allows the induction of hematopoietic stem/progenitor cell (HSPC) derived from human embryonic stem cells (hESCs) in vitro. The results showed that SMG facilitates hESCs differentiate to HSPC with more efficient induction of CD34+CD31+ hemogenic endothelium progenitors (HEPs) on day 4 and CD34+CD43+ HSPC on day 7, and these cells shows an increased generation of functional hematopoietic cells in colony-forming unit assay when compared with normal gravity (NG) conditions. Additionally, we found that SMG significantly increased the total number of cells on day 4 and day 7 which formed more 3D cell clusters. Transcriptome analysis of cells identified thousands of differentially expressed genes (DEGs) between NG and SMG. DEGs down-regulated were enriched in the axonogenesis, positive regulation of cell adhesion, cell adhesion molecule and axon guidance, while SMG resulted in the up-regulation of genes were functionally associated with DNA replication, cell cycle, PI3K-Akt signaling pathway and tumorigenesis. Interestingly, some key gene terms were enriched in SMG, like hypoxia and ECM receptor interaction. Moreover, HSPC obtained from SMG culture conditions had a robust ability of proliferation in vitro. The proliferated cells also had the ability to form erythroid, granulocyte and monocyte/macrophage colonies, and can be induced to generate macrophages and megakaryocytes. In summary, our data has shown a potent impact of microgravity on hematopoietic differentiation of hPSCs for the first time and reveals an underlying mechanism for the effect of SMG on hematopoiesis development.
Keywords: 3D clusters; hematopoietic differentiation; human embryonic stem cells; microgravity; transcriptome analysis.
Copyright © 2022 Ma, Xiong, Han, Zhang, Cao, Wang, Zhao, Duan, Zhang and Lei.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

