Matrin3: Disorder and ALS Pathogenesis
- PMID: 35083279
- PMCID: PMC8784776
- DOI: 10.3389/fmolb.2021.794646
Matrin3: Disorder and ALS Pathogenesis
Abstract
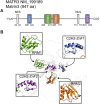
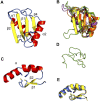
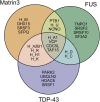
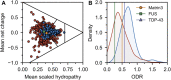
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder characterized by the degeneration of both upper and lower motor neurons in the brain and spinal cord. ALS is associated with protein misfolding and inclusion formation involving RNA-binding proteins, including TAR DNA-binding protein (TDP-43) and fused in sarcoma (FUS). The 125-kDa Matrin3 is a highly conserved nuclear DNA/RNA-binding protein that is implicated in many cellular processes, including binding and stabilizing mRNA, regulating mRNA nuclear export, modulating alternative splicing, and managing chromosomal distribution. Mutations in MATR3, the gene encoding Matrin3, have been identified as causal in familial ALS (fALS). Matrin3 lacks a prion-like domain that characterizes many other ALS-associated RNA-binding proteins, including TDP-43 and FUS, however, our bioinformatics analyses and preliminary studies document that Matrin3 contains long intrinsically disordered regions that may facilitate promiscuous interactions with many proteins and may contribute to its misfolding. In addition, these disordered regions in Matrin3 undergo numerous post-translational modifications, including phosphorylation, ubiquitination and acetylation that modulate the function and misfolding of the protein. Here we discuss the disordered nature of Matrin3 and review the factors that may promote its misfolding and aggregation, two elements that might explain its role in ALS pathogenesis.
Keywords: ALS; Matrin3; intrinsically disordered domains; protein misfolding; proteinopathy.
Copyright © 2022 Salem, Wilson, Rutledge, Dilliott, Farhan, Choy and Duennwald.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
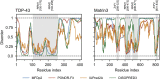
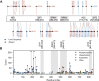
References
-
- Andrews J. A., Jackson C. E., Heiman-Patterson T. D., Bettica P., Brooks B. R., Pioro E. P. (2020). Real-world Evidence of Riluzole Effectiveness in Treating Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Frontotemporal Degeneration 21, 509–518. 10.1080/21678421.2020.1771734 - DOI - PubMed
-
- Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., et al. (2006). TDP-43 Is a Component of Ubiquitin-Positive Tau-Negative Inclusions in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Biochem. Biophysical Res. Commun. 351, 602–611. 10.1016/j.bbrc.2006.10.093 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

