Phage Annotation Guide: Guidelines for Assembly and High-Quality Annotation
- PMID: 35083439
- PMCID: PMC8785237
- DOI: 10.1089/phage.2021.0013
Phage Annotation Guide: Guidelines for Assembly and High-Quality Annotation
Abstract
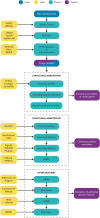
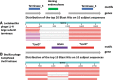
All sequencing projects of bacteriophages (phages) should seek to report an accurate and comprehensive annotation of their genomes. This article defines 14 questions for those new to phage genomics that should be addressed before submitting a genome sequence to the International Nucleotide Sequence Database Collaboration or writing a publication.
Keywords: annotation guide; bacteriophages; genome annotation; genomics; phage taxonomy; phages.
© Dann Turner et al. 2022; Published by Mary Ann Liebert, Inc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Holtappels D, Fortuna K, Lavigne R, et al. . The future of phage biocontrol in integrated plant protection for sustainable crop production. Curr Opin Biotechnol. 2021;68:60–71. - PubMed
-
- De Smet J, Hendrix H, Blasdel BG, et al. . Pseudomonas predators: Understanding and exploiting phage–host interactions. Nat Rev Microbiol. 2017;15(9):517–530. - PubMed
-
- Moreno Switt AI, Sulakvelidze A, Wiedmann M, et al. Salmonella phages and prophages: Genomics, taxonomy, and applied aspects. In: Schatten H, Eisenstark A; eds. Salmonella: Methods and Protocols, Methods in Molecular Biology: Volume 1225. New York: Springer Science + Business Media; 2015: 237–287. - PubMed
-
- Christie GE, Kuzio HE, McShan J, et al. Prophage-induced changes in cellular cytochemistry and virulence. In: Hyman P, Abedon ST; eds. Bacteriophages in Health and Disease. CABI Press; 2012.
Grants and funding
LinkOut - more resources
Full Text Sources
