Clinical challenges of tissue preparation for spatial transcriptome
- PMID: 35083877
- PMCID: PMC8792118
- DOI: 10.1002/ctm2.669
Clinical challenges of tissue preparation for spatial transcriptome
Abstract
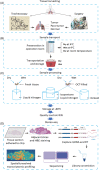
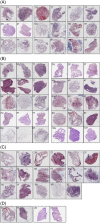
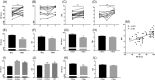
Spatial transcriptomics is considered as an important part of spatiotemporal molecular images to bridge molecular information with clinical images. Of those potentials and opportunities, the excellent quality of human sample preparation and handling will ensure the precise and reliable information generated from clinical spatial transcriptome. The present study aims at defining potential factors that might influence the quality of spatial transcriptomics in lung cancer, para-cancer, or normal tissues, pathological images of sections and the RNA integrity before spatial transcriptome sequencing. We categorised potential influencing factors from clinical aspects, including patient selection, pathological definition, surgical types, sample harvest, temporary preservation conditions and solutions, frozen approaches, transport and storage conditions and duration. We emphasis on the relationship between the combination of histological scores with RNA integrity number (RIN) and the unique molecular identifier (UMI), which is determines the quality of of spatial transcriptomics; however, we did not find significantly relevance between them. Our results showed that isolated times and dry conditions of sample are critical for the UMI and the quality of spatial transcriptomic samples. Thus, clinical procedures of sample preparation should be furthermore optimised and standardised as new standards of operation performance for clinical spatial transcriptome. Our data suggested that the temporary preservation time and condition of samples at operation room should be within 30 min and in 'dry' status. The direct cryo-preservation within OCT media for human lung sample is recommended. Thus, we believe that clinical spatial transcriptome will be a decisive approach and bridge in the development of spatiotemporal molecular images and provide new insights for understanding molecular mechanisms of diseases at multi-orientations.
Keywords: clinical challenge; critical factor; spatial transcriptomics; spatiotemporal molecular image; spatiotemporal molecular medicine.
© 2022 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg A, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78‐82. - PubMed
-
- Ao CSL, Mengnan C, Kailong M, et al. Large field of view‐spatially resolved transcriptomics at nanoscale resolution. bioRxiv. 2021. Jan 17.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
