Evaluating Eosinophilic Colitis as a Unique Disease Using Colonic Molecular Profiles: A Multi-Site Study
- PMID: 35085569
- PMCID: PMC9038694
- DOI: 10.1053/j.gastro.2022.01.022
Evaluating Eosinophilic Colitis as a Unique Disease Using Colonic Molecular Profiles: A Multi-Site Study
Abstract
Background & aims: Colonic eosinophilia, an enigmatic finding often referred to as eosinophilic colitis (EoC), is a poorly understood condition. Whether EoC is a distinct disease or a colonic manifestation of eosinophilic gastrointestinal diseases (EGIDs) or inflammatory bowel disease (IBD) is undetermined.
Methods: Subjects with EoC (n = 27) and controls (normal [NL, n = 20], Crohn's disease [CD, n = 14]) were enrolled across sites associated with the Consortium of Eosinophilic Gastrointestinal Disease Researchers. EoC was diagnosed as colonic eosinophilia (ascending ≥100, descending ≥85, sigmoid ≥65 eosinophils/high-power field) with related symptoms. Colon biopsies were subjected to RNA sequencing. Associations between gene expression and histologic features were analyzed with Spearman correlation; operational pathways and cellular constituents were computationally derived.
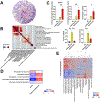
Results: We identified 987 differentially expressed genes (EoC transcriptome) between EoC and NL (>1.5-fold change, P < .05). Colonic eosinophil count correlated with 31% of EoC transcriptome, most notably with CCL11 and CLC (r = 0.78 and 0.77, P < .0001). Among EoC and other EGIDs, there was minimal transcriptomic overlap and minimal evidence of a strong allergic type 2 immune response in EoC compared with other EGIDs. Decreased cell cycle and increased apoptosis in EoC compared with NL were identified by functional enrichment analysis and immunostaining using Ki-67 and cleaved caspase-3. Pericryptal circumferential eosinophil collars were associated with the EoC transcriptome (P < .001). EoC transcriptome-based scores were reversible with disease remission and differentiated EoC from IBD, even after controlling for colonic eosinophil levels (P < .0001).
Conclusions: We established EoC transcriptomic profiles, identified mechanistic pathways, and integrated findings with parallel IBD and EGID data. These findings establish EoC as a distinct disease compared with other EGIDs and IBD, thereby providing a basis for improving diagnosis and treatment.
Keywords: Colitis; Eosinophil; Eosinophilic Colitis; Inflammatory Bowel Disease; Transcriptome.
Copyright © 2022. Published by Elsevier Inc.
Figures
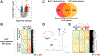



Comment in
-
Eosinophilic colitis - a distinct clinical entity?Nat Rev Gastroenterol Hepatol. 2022 Mar;19(3):148. doi: 10.1038/s41575-022-00583-0. Nat Rev Gastroenterol Hepatol. 2022. PMID: 35105954 No abstract available.
References
-
- Rothenberg ME. Eosinophilic gastrointestinal disorders (EGID). J Allergy Clin Immunol 2004;113:11–28. - PubMed
-
- Mansoor E, Saleh MA, Cooper GS. Prevalence of Eosinophilic Gastroenteritis and Colitis in a Population-Based Study, From 2012 to 2017. Clin Gastroenterol Hepatol 2017;15:1733–1741. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

