IRF8 is a transcriptional activator of CD37 expression in diffuse large B-cell lymphoma
- PMID: 35086136
- PMCID: PMC9006271
- DOI: 10.1182/bloodadvances.2021004366
IRF8 is a transcriptional activator of CD37 expression in diffuse large B-cell lymphoma
Abstract
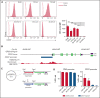
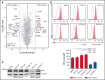
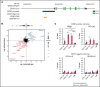
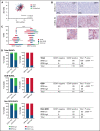
Diffuse large B-cell lymphoma (DLBCL) represents the most common form of non-Hodgkin lymphoma (NHL) that is still incurable in a large fraction of patients. Tetraspanin CD37 is highly expressed on mature B lymphocytes, and multiple CD37-targeting therapies are under clinical development for NHL. However, CD37 expression is nondetectable in ∼50% of DLBCL patients, which correlates with inferior treatment outcome, but the underlying mechanisms for differential CD37 expression in DLBCL are still unknown. Here, we investigated the regulation of the CD37 gene in human DLBCL at the (epi-)genetic and transcriptional level. No differences were observed in DNA methylation within the CD37 promoter region between CD37-positive and CD37-negative primary DLBCL patient samples. On the contrary, CD37-negative DLBCL cells specifically lacked CD37 promoter activity, suggesting differential regulation of CD37 gene expression. Using an unbiased quantitative proteomic approach, we identified transcription factor IRF8 to be significantly higher expressed in nuclear extracts of CD37-positive as compared with CD37-negative DLBCL. Direct binding of IRF8 to the CD37 promoter region was confirmed by DNA pulldown assay combined with mass spectrometry and targeted chromatin immunoprecipitation (ChIP). Functional analysis indicated that IRF8 overexpression enhanced CD37 protein expression, while CRISPR/Cas9 knockout of IRF8 decreased CD37 levels in DLBCL cell lines. Immunohistochemical analysis in a large cohort of primary DLBCL (n = 206) revealed a significant correlation of IRF8 expression with detectable CD37 levels. Together, this study provides new insight into the molecular mechanisms underlying differential CD37 expression in human DLBCL and reveals IRF8 as a transcriptional regulator of CD37 in B-cell lymphoma.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures

References
-
- Martelli M, Ferreri AJM, Agostinelli C, Di Rocco A, Pfreundschuh M, Pileri SA. Diffuse large B-cell lymphoma. Crit Rev Oncol Hematol. 2013;87(2):146-171. - PubMed
-
- Sarkozy C, Sehn LH. Management of relapsed/refractory DLBCL. Best Pract Res Clin Haematol. 2018;31(3):209-216. - PubMed
-
- Barrena S, Almeida J, Yunta M, et al. . Aberrant expression of tetraspanin molecules in B-cell chronic lymphoproliferative disorders and its correlation with normal B-cell maturation. Leukemia. 2005;19(8):1376-1383. - PubMed
-
- de Winde CM, Elfrink S, van Spriel AB. Novel Insights into membrane targeting of B cell lymphoma. Trends Cancer. 2017;3(6):442-453. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

