Replication of Dengue Virus in K562-Megakaryocytes Induces Suppression in the Accumulation of Reactive Oxygen Species
- PMID: 35087488
- PMCID: PMC8787197
- DOI: 10.3389/fmicb.2021.784070
Replication of Dengue Virus in K562-Megakaryocytes Induces Suppression in the Accumulation of Reactive Oxygen Species
Abstract
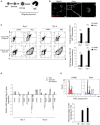
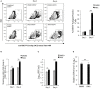
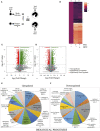
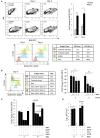
Dengue virus can infect human megakaryocytes leading to decreased platelet biogenesis. In this article, we report a study of Dengue replication in human K562 cells undergoing PMA-induced differentiation into megakaryocytes. PMA-induced differentiation in these cells recapitulates steps of megakaryopoiesis including gene activation, expression of CD41/61 and CD61 platelet surface markers and accumulation of intracellular reactive oxygen species (ROS). Our results show differentiating megakaryocyte cells to support higher viral replication without any apparent increase in virus entry. Further, Dengue replication suppresses the accumulation of ROS in differentiating cells, probably by only augmenting the activity of the transcription factor NFE2L2 without influencing the expression of the coding gene. Interestingly pharmacological modulation of NFE2L2 activity showed a simultaneous but opposite effect on intracellular ROS and virus replication suggesting the former to have an inhibitory effect on the later. Also cells that differentiated while supporting intracellular virus replication showed reduced level of surface markers compared to uninfected differentiated cells.
Keywords: Dengue virus replication; NFE2L2; flavivirus; megakaryopoiesis; reactive oxygen species.
Copyright © 2022 Kaur, Rawat, Sood, Periwal, Rathore, Kumar, Kumar and Bhattacharyya.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Dengue virus infection impedes megakaryopoiesis in MEG-01 cells where the virus envelope protein interacts with the transcription factor TAL-1.Sci Rep. 2020 Nov 11;10(1):19587. doi: 10.1038/s41598-020-76350-5. Sci Rep. 2020. PMID: 33177556 Free PMC article.
-
Effects of radiation on the maturation of megakaryocytes.J Radiat Res. 2013 May;54(3):447-52. doi: 10.1093/jrr/rrs127. Epub 2013 Jan 7. J Radiat Res. 2013. PMID: 23297317 Free PMC article.
-
Synergistic effect of hydrogen peroxide on polyploidization during the megakaryocytic differentiation of K562 leukemia cells by PMA.Exp Cell Res. 2013 Aug 15;319(14):2205-15. doi: 10.1016/j.yexcr.2013.06.002. Epub 2013 Jun 13. Exp Cell Res. 2013. PMID: 23770036 Free PMC article.
-
Molecular control of megakaryopoiesis and thrombopoiesis.Int J Hematol. 2002 Jun;75(5):473-83. doi: 10.1007/BF02982109. Int J Hematol. 2002. PMID: 12095146 Review.
-
Reactive oxygen species as potential antiviral targets.Rev Med Virol. 2022 Jan;32(1):e2240. doi: 10.1002/rmv.2240. Epub 2021 May 4. Rev Med Virol. 2022. PMID: 33949029 Review.
Cited by
-
Thrombocytopenia in dengue infection: mechanisms and a potential application.Expert Rev Mol Med. 2024 Oct 14;26:e26. doi: 10.1017/erm.2024.18. Expert Rev Mol Med. 2024. PMID: 39397710 Free PMC article. Review.
-
The up-regulation of TGF-β1 by miRNA-132-3p/WT1 is involved in inducing leukemia cells to differentiate into macrophages.PLoS One. 2025 May 6;20(5):e0306150. doi: 10.1371/journal.pone.0306150. eCollection 2025. PLoS One. 2025. PMID: 40327646 Free PMC article.
-
Targeting TLR2/Rac1/cdc42/JNK Pathway to Reveal That Ruxolitinib Promotes Thrombocytopoiesis.Int J Mol Sci. 2022 Dec 17;23(24):16137. doi: 10.3390/ijms232416137. Int J Mol Sci. 2022. PMID: 36555781 Free PMC article.
-
Dengue virus modulates critical cell cycle regulatory proteins in human megakaryocyte cells.Sci Rep. 2025 May 30;15(1):19016. doi: 10.1038/s41598-025-02640-5. Sci Rep. 2025. PMID: 40447783 Free PMC article.
-
Depletion of Bone Marrow Hematopoietic Cells in Ebolavirus-Infected Rhesus Macaques: A Possible Cause of Hematologic Abnormalities in Ebolavirus Disease.Am J Pathol. 2023 Dec;193(12):2031-2046. doi: 10.1016/j.ajpath.2023.08.010. Epub 2023 Sep 7. Am J Pathol. 2023. PMID: 37689386 Free PMC article.
References
-
- Cecchetti L., Tolley N. D., Michetti N., Bury L., Weyrich A. S., Gresele P. (2011). Megakaryocytes differentially sort mRNAs for matrix metalloproteinases and their inhibitors into platelets: a mechanism for regulating synthetic events. Blood 118, 1903–1911. 10.1182/blood-2010-12-324517 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

