GAL08, an Uncultivated Group of Acidobacteria, Is a Dominant Bacterial Clade in a Neutral Hot Spring
- PMID: 35087491
- PMCID: PMC8787282
- DOI: 10.3389/fmicb.2021.787651
GAL08, an Uncultivated Group of Acidobacteria, Is a Dominant Bacterial Clade in a Neutral Hot Spring
Abstract
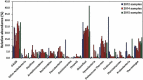
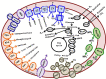
GAL08 are bacteria belonging to an uncultivated phylogenetic cluster within the phylum Acidobacteria. We detected a natural population of the GAL08 clade in sediment from a pH-neutral hot spring located in British Columbia, Canada. To shed light on the abundance and genomic potential of this clade, we collected and analyzed hot spring sediment samples over a temperature range of 24.2-79.8°C. Illumina sequencing of 16S rRNA gene amplicons and qPCR using a primer set developed specifically to detect the GAL08 16S rRNA gene revealed that absolute and relative abundances of GAL08 peaked at 65°C along three temperature gradients. Analysis of sediment collected over multiple years and locations revealed that the GAL08 group was consistently a dominant clade, comprising up to 29.2% of the microbial community based on relative read abundance and up to 4.7 × 105 16S rRNA gene copy numbers per gram of sediment based on qPCR. Using a medium quality threshold, 25 single amplified genomes (SAGs) representing these bacteria were generated from samples taken at 65 and 77°C, and seven metagenome-assembled genomes (MAGs) were reconstructed from samples collected at 45-77°C. Based on average nucleotide identity (ANI), these SAGs and MAGs represented three separate species, with an estimated average genome size of 3.17 Mb and GC content of 62.8%. Phylogenetic trees constructed from 16S rRNA gene sequences and a set of 56 concatenated phylogenetic marker genes both placed the three GAL08 bacteria as a distinct subgroup of the phylum Acidobacteria, representing a candidate order (Ca. Frugalibacteriales) within the class Blastocatellia. Metabolic reconstructions from genome data predicted a heterotrophic metabolism, with potential capability for aerobic respiration, as well as incomplete denitrification and fermentation. In laboratory cultivation efforts, GAL08 counts based on qPCR declined rapidly under atmospheric levels of oxygen but increased slightly at 1% (v/v) O2, suggesting a microaerophilic lifestyle.
Keywords: Acidobacteria; Acidobacteriota; GAL08; hot spring; single-cell genomics; thermophile; uncultivated bacterium.
Copyright © 2022 Ruhl, Sheremet, Furgason, Krause, Bowers, Jarett, Tran, Grasby, Woyke and Dunfield.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Ackerman G. G. (2006). Biogeochemical gradients and energetics in geothermal systems of Yellowstone National Park. Bozeman: Master’s dissertation, Montana State University.
-
- Amann R. I. (1995). In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes. Mole. Microb. Ecol. Man. 1995 1–15. 10.1007/978-94-011-0351-0_23 - DOI
-
- Baker B. J., Dick G. J. (2013). Omic approaches in microbial ecology: Charting the unknown. Microbe 8 353–360. 10.1128/microbe.8.353.1 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

