Setting-Up a Rapid SARS-CoV-2 Genome Assessment by Next-Generation Sequencing in an Academic Hospital Center (LPCE, Louis Pasteur Hospital, Nice, France)
- PMID: 35087842
- PMCID: PMC8787061
- DOI: 10.3389/fmed.2021.730577
Setting-Up a Rapid SARS-CoV-2 Genome Assessment by Next-Generation Sequencing in an Academic Hospital Center (LPCE, Louis Pasteur Hospital, Nice, France)
Abstract
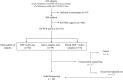
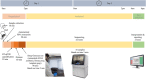
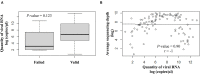
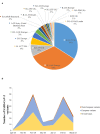
Introduction: Aside from the reverse transcription-PCR tests for the diagnosis of the COVID-19 in routine clinical care and population-scale screening, there is an urgent need to increase the number and the efficiency for full viral genome sequencing to detect the variants of SARS-CoV-2. SARS-CoV-2 variants assessment should be easily, rapidly, and routinely available in any academic hospital. Materials and Methods: SARS-CoV-2 full genome sequencing was performed retrospectively in a single laboratory (LPCE, Louis Pasteur Hospital, Nice, France) in 103 SARS-CoV-2 positive individuals. An automated workflow used the Ion Ampliseq SARS-CoV-2 panel on the Genexus Sequencer. The analyses were made from nasopharyngeal swab (NSP) (n = 64) and/or saliva (n = 39) samples. All samples were collected in the metropolitan area of the Nice city (France) from September 2020 to March 2021. Results: The mean turnaround time between RNA extraction and result reports was 30 h for each run of 15 samples. A strong correlation was noted for the results obtained between NSP and saliva paired samples, regardless of low viral load and high (>28) Ct values. After repeated sequencing runs, complete failure of obtaining a valid sequencing result was observed in 4% of samples. Besides the European strain (B.1.160), various variants were identified, including one variant of concern (B.1.1.7), and different variants under monitoring. Discussion: Our data highlight the current feasibility of developing the SARS-CoV-2 next-generation sequencing approach in a single hospital center. Moreover, these data showed that using the Ion Ampliseq SARS-CoV-2 Assay, the SARS-CoV-2 genome sequencing is rapid and efficient not only in NSP but also in saliva samples with a low viral load. The advantages and limitations of this setup are discussed.
Keywords: COVID-19; Genexus; SARS-CoV-2; next-generation sequencing (NGS); variants.
Copyright © 2022 Hofman, Bordone, Chamorey, Benzaquen, Schiappa, Lespinet-Fabre, Lanteri, Brest, Mograbi, Maniel, Tanga, Allegra, Salah, Fayada, Boutros, Leroy, Heeke, Hofman, Marquette and Ilié.
Conflict of interest statement
PH received honoraria from Thermo Fisher Scientific for participating in scientific meetings, outside this present work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

