Chromosome-Level Genome Assembly of a Human Fungal Pathogen Reveals Synteny among Geographically Distinct Species
- PMID: 35089059
- PMCID: PMC8725592
- DOI: 10.1128/mbio.02574-21
Chromosome-Level Genome Assembly of a Human Fungal Pathogen Reveals Synteny among Geographically Distinct Species
Abstract
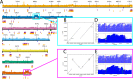
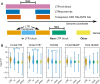
Histoplasma capsulatum, a dimorphic fungal pathogen, is the most common cause of fungal respiratory infections in immunocompetent hosts. Histoplasma is endemic in the Ohio and Mississippi River Valleys in the United States and is also distributed worldwide. Previous studies have revealed at least eight clades, each specific to a geographic location: North American classes 1 and 2 (NAm 1 and NAm 2), Latin American groups A and B (LAm A and LAm B), Eurasian, Netherlands, Australian and African, and an additional distinct lineage (H81) comprised of Panamanian isolates. Previously assembled Histoplasma genomes are highly fragmented, with the highly repetitive G217B (NAm 2) strain, which has been used for most whole-genome-scale transcriptome studies, assembled into over 250 contigs. In this study, we set out to fully assemble the repeat regions and characterize the large-scale genome architecture of Histoplasma species. We resequenced five Histoplasma strains (WU24 [NAm 1], G217B [NAm 2], H88 [African], G186AR [Panama], and G184AR [Panama]) using Oxford Nanopore Technologies long-read sequencing technology. Here, we report chromosomal-level assemblies for all five strains, which exhibit extensive synteny among the geographically distant Histoplasma isolates. The new assemblies revealed that RYP2, a major regulator of morphology and virulence, is duplicated in G186AR. In addition, we mapped previously generated transcriptome data sets onto the newly assembled chromosomes. Our analyses revealed that the expression of transposons and transposon-embedded genes are upregulated in yeast phase compared to mycelial phase in the G217B and H88 strains. This study provides an important resource for fungal researchers and further highlights the importance of chromosomal-level assemblies in analyzing high-throughput data sets. IMPORTANCE Histoplasma species are dimorphic fungi causing significant morbidity and mortality worldwide. These fungi grow as mold in the soil and as budding yeast within the human host. Histoplasma can be isolated from soil in diverse regions, including North America, South America, Africa, and Europe. Phylogenetically distinct species of Histoplasma have been isolated and sequenced. However, for the commonly used strains, genome assemblies have been fragmented, leading to underutilization of genome-scale data. This study provides chromosome-level assemblies of the commonly used Histoplasma strains using long-read sequencing technology. Comparative analysis of these genomes shows largely conserved gene order within the chromosomes. Mapping existing transcriptome data on these new assemblies reveals clustering of transcriptionally coregulated genes. The results of this study highlight the importance of obtaining chromosome-level assemblies in understanding the biology of human fungal pathogens.
Keywords: Histoplasma capsulatum; genome assembly; long-read sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Comparison of phylogenetically distinct Histoplasma strains reveals evolutionarily divergent virulence strategies.mBio. 2014 Jul 1;5(4):e01376-14. doi: 10.1128/mBio.01376-14. mBio. 2014. PMID: 24987093 Free PMC article.
-
Worldwide Phylogenetic Distributions and Population Dynamics of the Genus Histoplasma.PLoS Negl Trop Dis. 2016 Jun 1;10(6):e0004732. doi: 10.1371/journal.pntd.0004732. eCollection 2016 Jun. PLoS Negl Trop Dis. 2016. PMID: 27248851 Free PMC article.
-
Experimental annotation of the human pathogen Histoplasma capsulatum transcribed regions using high-resolution tiling arrays.BMC Microbiol. 2011 Sep 29;11:216. doi: 10.1186/1471-2180-11-216. BMC Microbiol. 2011. PMID: 21958208 Free PMC article.
-
[Intraspecific chromosomal variability in human pathogenic fungi, especially in Histoplasma capsulatum].Rev Iberoam Micol. 2004 Dec;21(4):168-76. Rev Iberoam Micol. 2004. PMID: 15709795 Review. Spanish.
-
Unraveling the secrets of Histoplasma capsulatum. A model to study morphogenic adaptation during parasite host/host interaction.Verh K Acad Geneeskd Belg. 1995;57(2):133-56. Verh K Acad Geneeskd Belg. 1995. PMID: 7571855 Review.
Cited by
-
How many missed abortions are caused by embryonic chromosomal abnormalities and what are their risk factors?Front Genet. 2023 Jan 4;13:1058261. doi: 10.3389/fgene.2022.1058261. eCollection 2022. Front Genet. 2023. PMID: 36685814 Free PMC article.
-
Genetic Diversity of Human Fungal Pathogens.Curr Clin Microbiol Rep. 2023 Jun;10(2):17-28. doi: 10.1007/s40588-023-00188-4. Epub 2023 Feb 14. Curr Clin Microbiol Rep. 2023. PMID: 37388463 Free PMC article.
-
Chromosome-level assemblies from diverse clades reveal limited structural and gene content variation in the genome of Candida glabrata.BMC Biol. 2022 Oct 8;20(1):226. doi: 10.1186/s12915-022-01412-1. BMC Biol. 2022. PMID: 36209154 Free PMC article.
-
An Indian lineage of Histoplasma with strong signatures of differentiation and selection.Fungal Genet Biol. 2022 Jan;158:103654. doi: 10.1016/j.fgb.2021.103654. Epub 2021 Dec 21. Fungal Genet Biol. 2022. PMID: 34942368 Free PMC article.
-
Datasets of fungal diversity and pseudo-chromosomal genomes of mangrove rhizosphere soil in China.Sci Data. 2024 Aug 20;11(1):901. doi: 10.1038/s41597-024-03748-5. Sci Data. 2024. PMID: 39164251 Free PMC article.
References
-
- Damasceno-Escoura AH, Mora DJ, Cardeal AC, Berto-Nascimento JC, Etchebehere RM, de Meneses ACO, Adad SJ, Micheletti AMR, Silva-Vergara ML. 2020. Histoplasmosis in HIV-infected patients: epidemiological, clinical and necropsy data from a Brazilian teaching hospital. Mycopathologia 185:339–346. doi:10.1007/s11046-020-00435-y. - DOI - PubMed
-
- Kasuga T, White TJ, Koenig G, Mcewen J, Restrepo A, Castañeda E, Da Silva Lacaz CDA, Heins-Vaccari EM, De Freitas RS, Zancopé-Oliveira RM, Qin Z, Negroni R, Carter DA, Mikami Y, Tamura M, Taylor ML, Miller GF, Poonwan N, Taylor JW. 2003. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol Ecol 12:3383–3401. doi:10.1046/j.1365-294x.2003.01995.x. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
