PixR, a Novel Activator of Conjugative Transfer of IncX4 Resistance Plasmids, Mitigates the Fitness Cost of mcr-1 Carriage in Escherichia coli
- PMID: 35089067
- PMCID: PMC8725589
- DOI: 10.1128/mbio.03209-21
PixR, a Novel Activator of Conjugative Transfer of IncX4 Resistance Plasmids, Mitigates the Fitness Cost of mcr-1 Carriage in Escherichia coli
Abstract
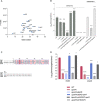
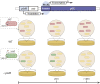
The emergence of the plasmid-borne colistin resistance gene mcr-1 threatens public health. IncX4-type plasmids are one of the most epidemiologically successful vehicles for spreading mcr-1 worldwide. Since MCR-1 is known for imposing a fitness cost to its host bacterium, the successful spread of mcr-1-bearing plasmids might be linked to high conjugation frequency, which would enhance the maintenance of the plasmid in the host without antibiotic selection. However, the mechanism of IncX4 plasmid conjugation remains unclear. In this study, we used high-density transposon mutagenesis to identify factors required for IncX4 plasmid transfer. Eighteen essential transfer genes were identified, including five with annotations unrelated to conjugation. Cappable-seq, transcriptome sequencing (RNA-seq), electrophoretic mobility shift assay, and β-galactosidase assay confirmed that a novel transcriptional regulator gene, pixR, directly regulates the transfer of IncX4 plasmids by binding the promoter of 13 essential transfer genes to increase their transcription. PixR is not active under nonmating conditions, while the expression of the pixR, pilX3-4, and pilX11 genes increased 3- to 6-fold upon contact with recipient Escherichia coli C600. Plasmid invasion and coculture competition assays revealed the essentiality of pixR for spreading and persistence of mcr-1-bearing IncX4 plasmids in bacterial populations. Effective conjugation is crucial for alleviating the fitness cost exerted by mcr-1 carriage. The existence of the IncX4-specific pixR gene increases plasmid transmissibility while promoting the invasion and persistence of mcr-1-bearing plasmids in bacterial populations, which helps explain their global prevalence. IMPORTANCE The spread of clinically relevant antibiotic resistance genes is often linked to the dissemination of epidemic plasmids. However, the underlying molecular mechanisms contributing to the successful spread of epidemic plasmids remain unclear. In this report, we shine a light on the transfer activation of IncX4 plasmids. We show how conjugation promotes the invasion and persistence of IncX4 plasmids within a bacterial population. The dissection of the regulatory network of conjugation helps explain the rapid spread of epidemic plasmids in nature. It also reveals potential targets for the development of conjugation inhibitors.
Keywords: IncX4; conjugation; fitness; mcr-1; plasmids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- World Health Organization. The ISSN register. https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance. Accessed 31 July 2020.
-
- Loftie-Eaton W, Yano H, Burleigh S, Simmons RS, Hughes JM, Rogers LM, Hunter SS, Settles ML, Forney LJ, Ponciano JM, Top EM. 2016. Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol Biol Evol 33:885–897. doi:10.1093/molbev/msv339. - DOI - PMC - PubMed
-
- Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi:10.1016/S1473-3099(17)30753-3. - DOI - PubMed
-
- Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi:10.1016/S1473-3099(15)00424-7. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
