Multimodal deep learning for biomedical data fusion: a review
- PMID: 35089332
- PMCID: PMC8921642
- DOI: 10.1093/bib/bbab569
Multimodal deep learning for biomedical data fusion: a review
Abstract
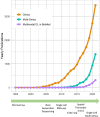
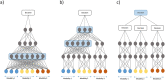
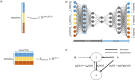
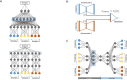
Biomedical data are becoming increasingly multimodal and thereby capture the underlying complex relationships among biological processes. Deep learning (DL)-based data fusion strategies are a popular approach for modeling these nonlinear relationships. Therefore, we review the current state-of-the-art of such methods and propose a detailed taxonomy that facilitates more informed choices of fusion strategies for biomedical applications, as well as research on novel methods. By doing so, we find that deep fusion strategies often outperform unimodal and shallow approaches. Additionally, the proposed subcategories of fusion strategies show different advantages and drawbacks. The review of current methods has shown that, especially for intermediate fusion strategies, joint representation learning is the preferred approach as it effectively models the complex interactions of different levels of biological organization. Finally, we note that gradual fusion, based on prior biological knowledge or on search strategies, is a promising future research path. Similarly, utilizing transfer learning might overcome sample size limitations of multimodal data sets. As these data sets become increasingly available, multimodal DL approaches present the opportunity to train holistic models that can learn the complex regulatory dynamics behind health and disease.
Keywords: data integration; deep neural networks; fusion strategies; multi-omics; multimodal machine learning; representation learning.
© The Author(s) 2022. Published by Oxford University Press.
Figures
References
-
- Ramachandram D, Taylor GW. Deep multimodal learning: a survey on recent advances and trends. IEEE Signal Process Mag 2017;34(6):96–108.
-
- Hall DL, Llinas J. An introduction to multisensor data fusion. Proc IEEE 1997;85(1):6–23.
-
- Durrant-Whyte HF. Sensor models and multisensor integration. Int J Robot Res 1988;7:97–113.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

