Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants
- PMID: 35090164
- PMCID: PMC8967717
- DOI: 10.1038/s41586-022-04466-x
Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants
Abstract
Omicron (B.1.1.529), the most heavily mutated SARS-CoV-2 variant so far, is highly resistant to neutralizing antibodies, raising concerns about the effectiveness of antibody therapies and vaccines1,2. Here we examined whether sera from individuals who received two or three doses of inactivated SARS-CoV-2 vaccine could neutralize authentic Omicron. The seroconversion rates of neutralizing antibodies were 3.3% (2 out of 60) and 95% (57 out of 60) for individuals who had received 2 and 3 doses of vaccine, respectively. For recipients of three vaccine doses, the geometric mean neutralization antibody titre for Omicron was 16.5-fold lower than for the ancestral virus (254). We isolated 323 human monoclonal antibodies derived from memory B cells in triple vaccinees, half of which recognized the receptor-binding domain, and showed that a subset (24 out of 163) potently neutralized all SARS-CoV-2 variants of concern, including Omicron. Therapeutic treatments with representative broadly neutralizing monoclonal antibodies were highly protective against infection of mice with SARS-CoV-2 Beta (B.1.351) and Omicron. Atomic structures of the Omicron spike protein in complex with three classes of antibodies that were active against all five variants of concern defined the binding and neutralizing determinants and revealed a key antibody escape site, G446S, that confers greater resistance to a class of antibodies that bind on the right shoulder of the receptor-binding domain by altering local conformation at the binding interface. Our results rationalize the use of three-dose immunization regimens and suggest that the fundamental epitopes revealed by these broadly ultrapotent antibodies are rational targets for a universal sarbecovirus vaccine.
© 2022. The Author(s).
Conflict of interest statement
Y.H., Lin Wang and M.L. are employees of Sinovac Biotech Ltd. Y.J., P.G. and Y.C. are employees of Acrobiosystems Inc. The other authors declare no competing interests.
Figures









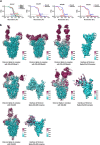





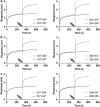

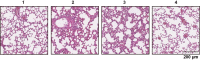
Comment in
-
A booster with SARS-CoV-2 vaccines: protection against Omicron infection.Signal Transduct Target Ther. 2022 Apr 5;7(1):115. doi: 10.1038/s41392-022-00973-5. Signal Transduct Target Ther. 2022. PMID: 35383164 Free PMC article. No abstract available.
References
-
- Carreno, J. M. et al. Activity of convalescent and vaccine serum against a B. 1.1. 529 variant SARS-CoV-2 isolate. Nature10.1038/s41586-022-04399-5 (2021).
-
- Cao, Y. R. et al. B. 1.1. 529 escapes the majority of SARS-CoV-2 neutralizing antibodies of diverse epitopes. Nature10.1038/s41586-021-04385-3 (2021).
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

