Multiattribute Glycan Identification and FDR Control for Glycoproteomics
- PMID: 35091091
- PMCID: PMC8933705
- DOI: 10.1016/j.mcpro.2022.100205
Multiattribute Glycan Identification and FDR Control for Glycoproteomics
Abstract
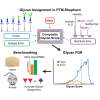
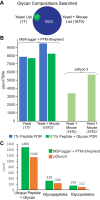
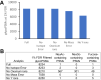
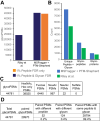
Rapidly improving methods for glycoproteomics have enabled increasingly large-scale analyses of complex glycopeptide samples, but annotating the resulting mass spectrometry data with high confidence remains a major bottleneck. We recently introduced a fast and sensitive glycoproteomics search method in our MSFragger search engine, which reports glycopeptides as a combination of a peptide sequence and the mass of the attached glycan. In samples with complex glycosylation patterns, converting this mass to a specific glycan composition is not straightforward; however, as many glycans have similar or identical masses. Here, we have developed a new method for determining the glycan composition of N-linked glycopeptides fragmented by collisional or hybrid activation that uses multiple sources of information from the spectrum, including observed glycan B-type (oxonium) and Y-type ions and mass and precursor monoisotopic selection errors to discriminate between possible glycan candidates. Combined with false discovery rate estimation for the glycan assignment, we show that this method is capable of specifically and sensitively identifying glycans in complex glycopeptide analyses and effectively controls the rate of false glycan assignments. The new method has been incorporated into the PTM-Shepherd modification analysis tool to work directly with the MSFragger glyco search in the FragPipe graphical user interface, providing a complete computational pipeline for annotation of N-glycopeptide spectra with false discovery rate control of both peptide and glycan components that is both sensitive and robust against false identifications.
Keywords: false discovery rate; glycoproteomics; glycosylation; mass spectrometry; software.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

