Application of liquid biopsy-based targeted capture sequencing analysis to improve the precision treatment of non-small cell lung cancer by tyrosine kinase inhibitors
- PMID: 35091437
- PMCID: PMC8804681
- DOI: 10.1136/bmjresp-2021-001154
Application of liquid biopsy-based targeted capture sequencing analysis to improve the precision treatment of non-small cell lung cancer by tyrosine kinase inhibitors
Abstract
Background: Targeted therapy of patients with non-small cell lung cancer (NSCLC) who harbour sensitising mutations by tyrosine kinase inhibitors (TKIs) has been found more effective than traditional chemotherapies. However, target genes status (eg, epidermal growth factor receptor (EGFR) TKIs sensitising and resistant mutations) need to be tested for choosing appropriate TKIs. This study is to investigate the performance of a liquid biopsy-based targeted capture sequencing assay on the molecular analysis of NSCLC.
Methods: Plasma samples from patients with NSCLC who showed resistance to the first/second-generation EGFR TKIs treatment were collected. The AVENIO ctDNA Expanded Kit is a 77 pan-cancer genes detection assay that was used for detecting EGFR TKIs resistance-associated gene mutations. Through comparison of the EGFR gene testing results from the Cobas EGFR Mutation Test v2, and UltraSEEK Lung Panel, the effectiveness of the targeted capture sequencing assay was verified.
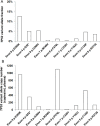
Results: A total of 24 plasma cell-free DNA (cfDNA) samples were tested by the targeted capture sequencing assay. 33.3% (8/24) cfDNA samples were positive for EGFR exon 20 p.T790M which leads to EGFR dependent TKIs resistance. 8.3% (2/24) and 4.2% (1/24) samples were positive for mesenchymal-epithelial transition gene amplification and B-Raf proto-oncogene, serine/threonine kinase exon 15 p.V600E mutations which lead to EGFR independent TKIs resistance. The median value of the p.T790M variant allele fraction and variant copy numbers was 2% and 36.10 copies/mL plasma, respectively. The next-generation sequencing test showed higher than 90% concordance with either MassArray or qPCR-based methods for detecting either EGFR TKIs sensitising or resistance mutations.
Conclusion: The targeted capture sequencing test can support comprehensive molecular analysis needed for TKIs treatment, which is promising to be clinically applied for the improved precision treatment of NSCLC.
Keywords: drug reactions; lung cancer; non-small cell lung cancer.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: II has received honorarium for consultation from AstraZeneca, Roche, Bayer. The other authors of this study declare that they have no competing interests.
Figures



References
-
- Canadian-Cancer-Statistics-Advisory-Committee . Canadian cancer statistics 2018. Canadian Cancer Society, 2018.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
