Oligonucleotide correction of an intronic TIMMDC1 variant in cells of patients with severe neurodegenerative disorder
- PMID: 35091571
- PMCID: PMC8799713
- DOI: 10.1038/s41525-021-00277-7
Oligonucleotide correction of an intronic TIMMDC1 variant in cells of patients with severe neurodegenerative disorder
Abstract
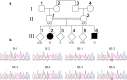
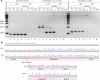
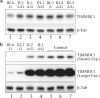
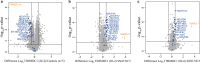
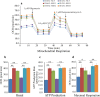
TIMMDC1 encodes the Translocase of Inner Mitochondrial Membrane Domain-Containing protein 1 (TIMMDC1) subunit of complex I of the electron transport chain responsible for ATP production. We studied a consanguineous family with two affected children, now deceased, who presented with failure to thrive in the early postnatal period, poor feeding, hypotonia, peripheral neuropathy and drug-resistant epilepsy. Genome sequencing data revealed a known, deep intronic pathogenic variant TIMMDC1 c.597-1340A>G, also present in gnomAD (~1/5000 frequency), that enhances aberrant splicing. Using RNA and protein analysis we show almost complete loss of TIMMDC1 protein and compromised mitochondrial complex I function. We have designed and applied two different splice-switching antisense oligonucleotides (SSO) to restore normal TIMMDC1 mRNA processing and protein levels in patients' cells. Quantitative proteomics and real-time metabolic analysis of mitochondrial function on patient fibroblasts treated with SSOs showed restoration of complex I subunit abundance and function. SSO-mediated therapy of this inevitably fatal TIMMDC1 neurologic disorder is an attractive possibility.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Kumar R, et al. Homozygous mutation of STXBP5L explains an autosomal recessive infantile-onset neurodegenerative disorder. Hum. Mol. Genet. 2015;24:2000–2010. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

