Depletion of mitochondrial methionine adenosyltransferase α1 triggers mitochondrial dysfunction in alcohol-associated liver disease
- PMID: 35091576
- PMCID: PMC8799735
- DOI: 10.1038/s41467-022-28201-2
Depletion of mitochondrial methionine adenosyltransferase α1 triggers mitochondrial dysfunction in alcohol-associated liver disease
Abstract
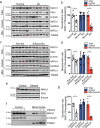
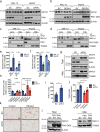
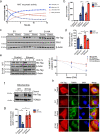
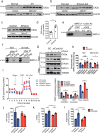
MATα1 catalyzes the synthesis of S-adenosylmethionine, the principal biological methyl donor. Lower MATα1 activity and mitochondrial dysfunction occur in alcohol-associated liver disease. Besides cytosol and nucleus, MATα1 also targets the mitochondria of hepatocytes to regulate their function. Here, we show that mitochondrial MATα1 is selectively depleted in alcohol-associated liver disease through a mechanism that involves the isomerase PIN1 and the kinase CK2. Alcohol activates CK2, which phosphorylates MATα1 at Ser114 facilitating interaction with PIN1, thereby inhibiting its mitochondrial localization. Blocking PIN1-MATα1 interaction increased mitochondrial MATα1 levels and protected against alcohol-induced mitochondrial dysfunction and fat accumulation. Normally, MATα1 interacts with mitochondrial proteins involved in TCA cycle, oxidative phosphorylation, and fatty acid β-oxidation. Preserving mitochondrial MATα1 content correlates with higher methylation and expression of mitochondrial proteins. Our study demonstrates a role of CK2 and PIN1 in reducing mitochondrial MATα1 content leading to mitochondrial dysfunction in alcohol-associated liver disease.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

